LncRNA-TUG1 as a potential diagnostic biomarker for coronary atherosclerotic heart disease
doi: 10.1515/fzm-2025-0025
-
Abstract:
Objectives Accumulating evidence suggests that people living in cold regions have a higher risk of developing coronary atherosclerotic heart disease (CHD). Long non-coding RNAs (lncRNAs) have been implicated in the pathogenesis and treatment of a variety of diseases. The present study aimed to investigate the serum level of lncRNA-taurine upregulated gene 1 (TUG1) in patients with CHD and assess its potential as a diagnostic biomarker. This study aimes to investigate the serum level of lncRNA-TUG1 in patients with CHD and assess its potential as a diagnostic biomarker. Methods The Gene Expression Omnibus (GEO) database was employed to identify the potential lncRNAs serving as biomarkers for CHD. To validate lncRNA-TUG1, 232 subjects were enrolled in both test and diagnostic cohorts. Serum lncRNA-TUG1 levels were measured by RT-qPCR. The association between lncRNA-TUG1 levels and CHD severity was analyzed using Pearson's correlation test. Diagnostic value was assessed by receiver operating characteristic (ROC) curve analysis and compared with established cardiac biomarkers. Results LncRNA-TUG1 was identified in the GEO database as a potential biomarker for CHD. Serum lncRNA-TUG1 levels were significantly higher in CHD patients compared with healthy controls and non-CHD patients. CRP levels also differed between CHD and non-CHD groups, while other biomarkers showed no significant differences. ROC curve analysis demonstrated that lncRNA-TUG1 could distinguish CHD from non-CHD patients, with an area under the curve (AUC) of 0.8916, which was higher than that of conventional biomarkers such as cTnI. At a cut-off value of 2.311, the sensitivity and specificity of lncRNA-TUG1 were 61.63% and 97.67%, respectively, surpassing the diagnostic performance of cTnI. Furthermore, lncRNA-TUG1 levels in CHD patients were positively correlated with SYNTAX scores from coronary angiography and increased with the severity of vascular stenosis. Conclusion Elevated serum lncRNA-TUG1 levels in CHD patients suggest that lncRNA-TUG1 may serve as a novel and valuable diagnostic biomarker for CHD, with potential utility in differentiating CHD from other cardiac diseases. -
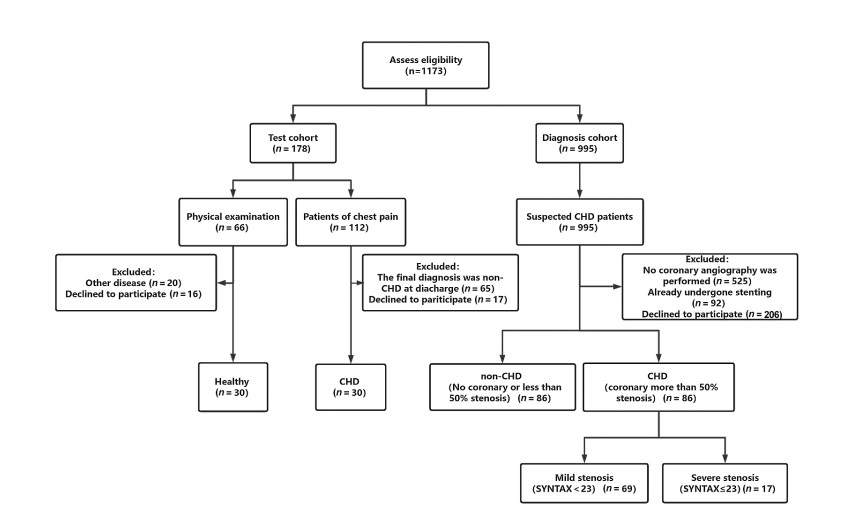
Figure 1. Flow diagram illustrating the identification and enrollment of study participants
From March 2019 to May 2023, patients admitted to the Department of Cardiology at the Fourth Affiliated Hospital of Harbin Medical University were screened according to inclusion and exclusion criteria. A total of 60 subjects were included in the test cohort (30 coronary atherosclerotic heart disease patients and 30 healthy controls). An additional 172 subjects were enrolled in the diagnostic cohort, comprising 86 CHD patients and 86 non-CHD patients confirmed by coronary angiography. CHD, coronary atherosclerotic heart disease.
Figure 2. Long non-coding RNA (lncRNA) screening for coronary atherosclerotic heart disease (CHD) using the Gene Expression Omnibus (GEO) database
(A) Volcano plot of differentially expressed genes in peripheral blood from CHD patients and healthy controls (GSE71226). (B) Volcano plot of differentially expressed genes in peripheral blood mononuclear cells from CHD patients before and after treatment with Tongmai Yangxin Pill (GSE142008). (C) Venn diagram showing 156 overlapping differentially expressed genes between GSE71226 and GSE142008 datasets, highlighting taurine upregulated gene 1 (TUG1). (D) Genomic location of TUG1, identified as a long non-coding RNA with potential diagnostic relevance in CHD.
Figure 3. Diagnostic ability of long non-coding RNA (lncRNA)-taurine upregulated gene 1 (TUG1) in the test cohort
(A) Serum lncRNA-TUG1 expression levels in patients with coronary atherosclerotic heart disease (CHD) compared with healthy controls. (B) Receiver operating characteristic (ROC) curve analysis demonstrating the diagnostic performance of lncRNA-TUG1 for distinguishing CHD patients from healthy individuals.
Figure 4. Analysis of long non-coding RNA (lncRNA)-taurine upregulated gene 1 (TUG1) in the diagnosis cohort
(A) Serum lncRNA-TUG1 levels in non-coronary atherosclerotic heart disease (non-CHD) patients and CHD patients. (B) Serum lncRNA-TUG1 levels in non-CHD patients, CHD patients with mild stenosis (SYNTAX score < 23), and CHD patients with severe stenosis (SYNTAX score ≥ 23). (C) Serum lncRNA-TUG1 levels in non-CHD patients, CHD patients with single-vessel stenosis, and CHD patients with multi-vessel stenosis.
Figure 5. Correlation analysis between long non-coding RNA (lncRNA)-taurine upregulated gene 1 (TUG1) expression and SYNTAX score in coronary atherosclerotic heart disease (CHD) patients
(A) Correlation between serum lncRNA-TUG1 levels and SYNTAX score in all CHD patients. (B) Correlation between lncRNA-TUG1 expression and SYNTAX score in CHD patients with mild stenosis (SYNTAX score < 23). (C) Correlation between lncRNA-TUG1 expression and SYNTAX score in CHD patients with severe stenosis (SYNTAX score ≥ 23).
Figure 6. Serum levels of conventional cardiac disease biomarkers in the diagnostic cohort
Box plots showing the distribution of (A) cTnI, (B) NT-proBNP, (C) HCY, (D) CRP, (E) CK, (F) CK-MB, (G) LDH, (H) HBDH, and (I) AST in patients with CHD compared with non-CHD patients. Among these biomarkers, only CRP was significantly elevated in CHD patients, while no significant differences were observed for the others. cTnI, cardiac troponin Ⅰ; NT-proBNP, N-terminal pro-B-type natriuretic peptide precursor; HCY, Homocysteine; CRP, C-reactive protein; CK, creatine kinase; LDH, lactate dehydrogenase; HBDH, alpha-hydroxybutyrate dehydrogenase; AST, aspartate aminotransferase; CHD, coronary atherosclerotic heart disease.
Figure 7. Diagnostic efficacy of lncRNA TUG1 for CHD
(A) Receiver operating characteristic (ROC) curves comparing the predictive value of lncRNA-TUG1 with conventional cardiac disease biomarkers (cTnI, NT-proBNP, CRP, HCY, CK, CK-MB, LDH, HBDH, and AST). (B-K) Dot plots showing the distribution of lncRNA-TUG1 values in non-CHD and CHD patients, compared with the distribution of conventional cardiac biomarkers across outcome groups. lnc-RNA, long non-coding RNA; TUG1, taurine upregulated gene 1; CHD, coronary atherosclerotic heart disease; cTnI, cardiac troponin Ⅰ; NT-proBNP, N-terminal pro-B-type natriuretic peptide precursor; HCY, Homocysteine; CRP, C-reactive protein; CK, creatine kinase; LDH, lactate dehydrogenase; HBDH, alpha-hydroxybutyrate dehydrogenase; AST, aspartate aminotransferase.
Table 1. Sequences of primers
Primers Forward (5'-3') Reverse LncRNA-TUG1 5'-TAGCAGTTCCCCAATCCTTG-3' 5'-CACAAATTCCCATCATTCCC-3' U6 5'-CTCGCTTCGGCAGCACATATACT-3' 5'-ACGCTTCACGAATTTGCGTGTC-3' Table 2. Clinical characteristics of each group
Clinical characteristics Test cohort Diagnosis cohort Healthy CHD P value Control CHD P value Age (year) 58 ± 7 66 ± 10 0.0055 69 ± 13 65 ± 10 0.0541 Gender (M/F) 30 (18/12) 30 (16/14) 0.6023 86 (52/34) 86 (59/27) 0.3390 SBP (mmHg) 120.5 ± 12.5 140.7 ± 17.3 0.0013 138.69 ± 22.16 137.61 ± 18.01 0.9592 DBP (mmHg) 73.5 ± 6.29 83.83 ± 10.0 0.0008 83.40 ± 14.28 82.70 ± 11.16 0.6870 Smoking (%) 10 (30.33) 6 (20.00) 0.2429 20 (23.53) 32 (37.21) 0.0673 Hypertension (%) 13 (43.33) 20 (66.67) 0.0693 45 (52.30) 45 (52.30) > 0.9999 Diabetes (%) 9 (30.00) 9 (30.00) > 0.9999 31 (36.05) 32 (37.21) > 0.9999 Hyperlipidemia (%) 16 (53.33) 25 (83.33) 0.0125 30 (34.88) 24 (27.91) 0.4115 LDL-C (mmol/L) 1.90 ± 0.99 2.28 ± 1.32 0.6691 2.79 ± 1.07 2.56 ± 0.88 0.2056 TG (mmol/L) 1.61 ± 0.49 3.65 ± 1.48 < 0.0001 1.64 ± 0.92 1.75 ± 0.99 0.2996 TC (mmol/L) 3.76 ± 1.34 3.60 ± 1.10 0.0319 4.16 ± 1.49 4.44 ± 1.16 0.1190 CHD, coronary atherosclerotic heart disease; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride. Table 3. Criterion values and coordinates of the receiver operating characteristic (ROC) curve for Long non-coding RNAs (lncRNAs)-taurine upregulated gene 1 (TUG1)
LncRNA-TUG1 Sensitivity 95%CI Specificity 95%CI > 0.217 100 95.720 to 100 1.163 0.060 to 6.296 > 1.548 82.560 73.200 to 89.140 73.260 63.050 to 81.470 > 2.311 61.630 51.060 to 71.200 97.670 91.910 to 99.590 > 8.110 1.163 0.060 to 6.296 100 95.720 to 100 -
[1] Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet, 2012; 380(9859): 2095-2128. doi: 10.1016/S0140-6736(12)61728-0 [2] Khera A V, Emdin C A, Drake I, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med, 2016; 375(24): 2349-2358. doi: 10.1056/NEJMoa1605086 [3] Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med, 2018; 22(12): 5768-5775. doi: 10.1111/jcmm.13866 [4] Liao J, Wang J, Liu Y, et al. Transcriptome sequencing of lncRNA, miRNA, mRNA and interaction network constructing in coronary heart disease. BMC Med Genomics, 2019; 12(1): 124. doi: 10.1186/s12920-019-0570-z [5] Schmitz S U, Grote P, Herrmann B G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci, 2016; 73(13): 2491-2509. doi: 10.1007/s00018-016-2174-5 [6] Sun J, Hu J, Wang G, et al. LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res, 2018; 37(1): 106. doi: 10.1186/s13046-018-0771-x [7] Han Y, Liu C, Lei M, et al. LncRNA TUG1 was upregulated in osteoporosis and regulates the proliferation and apoptosis of osteoclasts. J Orthop Surg Res, 2019; 14(1): 416. doi: 10.1186/s13018-019-1430-4 [8] Qiu N, Xu X, He Y. LncRNA TUG1 alleviates sepsis-induced acute lung injury by targeting miR-34b-5p/GAB1. BMC Pulm Med, 2020; 20(1): 49. doi: 10.1186/s12890-020-1084-3 [9] Du H, Yang L, Zhang H, et al. LncRNA TUG1 silencing enhances proliferation and migration of ox-LDL-treated human umbilical vein endothelial cells and promotes atherosclerotic vascular injury repairing via the Runx2/ANPEP axis. Int J Cardiol, 2021; 338: 204-214. doi: 10.1016/j.ijcard.2021.05.014 [10] Su Q, Liu Y, Lv X W, et al. Inhibition of lncRNA TUG1 upregulates miR-142-3p to ameliorate myocardial injury during ischemia and reperfusion via targeting HMGB1- and Rac1-induced autophagy. J Mol Cell Cardiol, 2019; 133: 12-25. doi: 10.1016/j.yjmcc.2019.05.021 [11] Su Y, Sun Y, Tang Y, et al. Circulating miR-19b-3p as a novel prognostic biomarker for acute heart failure. J Am Heart Assoc, 2021; 10(20): e022304. doi: 10.1161/JAHA.121.022304 [12] Scanlon P J, Faxon D P, Audet A M, et al. ACC/AHA guidelines for coronary angiography: Executive summary and recommendations. A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Committee on Coronary Angiography) developed in collaboration with the Society for Cardiac Angiography and Interventions. Circulation, 1999; 99(17): 2345-2357. doi: 10.1161/01.CIR.99.17.2345 [13] Malhotra R K, Indrayan A. A simple nomogram for sample size for estimating sensitivity and specificity of medical tests. Indian J Ophthalmol, 2010; 58(6): 519-522. doi: 10.4103/0301-4738.71699 [14] Lih O S, Jahmunah V, San T R, et al. Comprehensive electrocardiographic diagnosis based on deep learning. Artif Intell Med, 2020; 103: 101789. doi: 10.1016/j.artmed.2019.101789 [15] Ghadrdoost B, Haghjoo M, Firouzi A. Accuracy of cardiogoniometry compared with electrocardiography in the diagnosis of coronary artery disease. Res Cardiovasc Med, 2015; 4(1): e25547. doi: 10.5812/cardiovascmed.25547 [16] Kim H M, Kim H L, Kim M A et al. Additional roles of diastolic parameters in the diagnosis of obstructive coronary artery disease. Coron Artery Dis, 2021; 32(2): 145-151. doi: 10.1097/MCA.0000000000000970 [17] Arbab-Zadeh A, Di Carli M F, Cerci R, et al. Accuracy of computed tomographic angiography and single-photon emission computed tomography-acquired myocardial perfusion imaging for the diagnosis of coronary artery disease. Circ Cardiovasc Imaging, 2015; 8(10): e003533. doi: 10.1161/CIRCIMAGING.115.003533 [18] Li H, Sun K, Zhao R, et al. Inflammatory biomarkers of coronary heart disease. Front Biosci (Schol Ed), 2018; 10: 185-196. doi: 10.2741/s508 [19] Nair M, Sandhu S S, Sharma A K. Cancer molecular markers: A guide to cancer detection and management. Semin Cancer Biol, 2018; 52(Pt 1): 39-55. doi: 10.1016/j.semcancer.2018.02.002 [20] Vegter E L, Van der Meer P, De Windt L J, et al. MicroRNAs in heart failure: From biomarker to target for therapy. Eur J Heart Fail, 2016; 18(5): 457-468. doi: 10.1002/ejhf.495 [21] Meng S, Zhou H, Feng Z, et al. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol Cancer, 2017; 16(1): 94. doi: 10.1186/s12943-017-0663-2 [22] Beermann J, Piccoli M T, Viereck J, et al. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev, 2016; 96(4): 1297-1325. doi: 10.1152/physrev.00041.2015 [23] Guo F, Tang C, Li Y et al. The interplay of LncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-kappaB signalling pathway. J Cell Mol Med, 2018; 22(10): 5062-5075. doi: 10.1111/jcmm.13790 [24] Zhu Y, Yang T, Duan J et al. MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging (Albany NY), 2019; 11(4): 1089-1109. doi: 10.18632/aging.101766 [25] Xiong G, Jiang X, Song T. The overexpression of lncRNA H19 as a diagnostic marker for coronary artery disease. Rev Assoc Med Bras (1992), 2019; 65(2): 110-117. doi: 10.1590/1806-9282.65.2.110 [26] Du H, Yang L, Zhang H, et al. LncRNA TUG1 silencing enhances proliferation and migration of ox-LDL-treated human umbilical vein endothelial cells and promotes atherosclerotic vascular injury repairing via the Runx2/ANPEP axis. Int J Cardiol, 2021; 338: 204-214. doi: 10.1016/j.ijcard.2021.05.014 [27] Tang Y, Hu J, Zhong Z, et al. Long noncoding RNA TUG1 promotes the function in ox-LDL-Treated HA-VSMCs via miR-141-3p/ROR2 axis. Cardiovasc Ther, 2020; 2020: 6758934. doi: 10.1155/2020/6758934 [28] Yang L, Li T. LncRNA TUG1 regulates ApoM to promote atherosclerosis progression through miR-92a/FXR1 axis. J Cell Mol Med, 2020; 24(15): 8836-8848. doi: 10.1111/jcmm.15521 [29] Fu D, Gao T, Liu M, et al. LncRNA TUG1 aggravates cardiomyocyte apoptosis and myocardial ischemia/reperfusion injury. Histol Histopathol, 2021; 36(12): 1261-1272. [30] You G, Long X, Song F, et al. Metformin activates the AMPK-mTOR pathway by modulating lncRNA TUG1 to induce autophagy and inhibit atherosclerosis. Drug Des Devel Ther, 2020; 14: 457-468. doi: 10.2147/DDDT.S233932 -


 投稿系统
投稿系统


 下载:
下载: