The function and effect on prognosis of ANXA2 in gastric cancer peritoneal metastasis patients in cold region
doi: 10.1515/fzm-2024-0024
-
Abstract:
Objective Heilongjiang Province is part of the northern cold areas of China, where gastric cancer is one of the most common gastrointestinal malignancies. Peritoneal metastases (PM) are the leading cause of mortality among patients. This study conducted bioinformatics and basic research on the gene ANXA2 (Annexin A2), which may influence the prognosis of patients. Methods Genome sequencing was performed on patients from Heilongjiang to identify potential genes impacting survival time. The function of ANXA2 in gastric cancer was analyzed using multiple bioinformatics databases, focusing on its pathways and mechanisms. ANXA2-knockout gastric cancer cell lines were constructed, and in vitro assays, including CCK-8, flow cytometry, scratch, and Transwell experiments, were conducted. A nude mouse tumorigenesis model was also developed to analyze in vivo effects. Results ANXA2 was found to be expressed at higher levels in gastric cancer tissue than in normal gastric tissue, and its mRNA levels were associated with short overall survival (OS). Enrichment analysis indicated that ANXA2 is primarily localized on the cell membrane and primarily influences the PI3K-AKT signaling pathway. Cytological experiments demonstrated that knockdown of ANXA2 suppresses the growth and migration of gastric carcinoma cells, an effect that was also observed in vivo. Conclusions ANXA2 is essential for gastric cancer growth and may represent a potential risk factor affecting the survival probability of patients in cold regions. -
Key words:
- Gastric cancer /
- ANXA2 /
- prognosis survival /
- bioinformatics /
- cold region
-
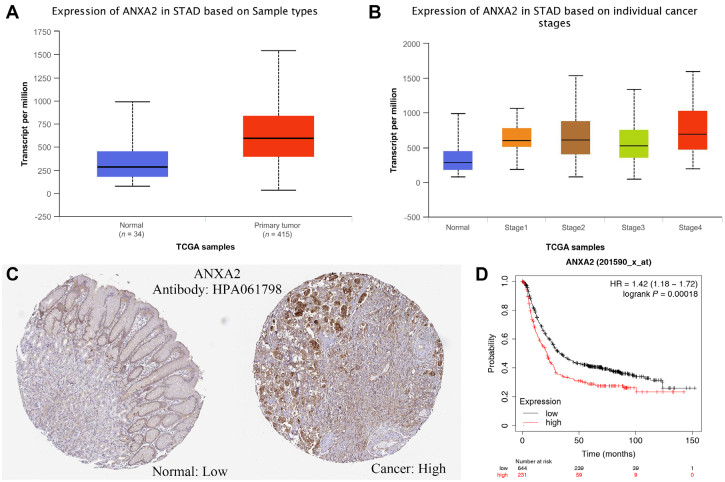
Figure 1. Bioinformatics results of ANXA2
(A) Changes of ANXA2 mRNA expression levels in gastric cancer and normal gastric tissues; (B) Expression levels of ANXA2 mRNA in the tissues of gastric cancer patients at different clinical stages; (C) Immunohistochemical analysis of ANXA2 protein expression in gastric cancer and normal tissues; (D) Relationship between ANXA2 mRNA levels and the survival of gastric cancer patients.
Figure 2. Enrichment results and relationship with PDL1 and ANXA2
(A-B) Enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) related to ANXA2; (C) The PI3K-AKT pathway in KEGG database; (D) Protein interaction network of ANXA2; (E) Relationship between mRNA expression of ANXA2 and PDL1; (F) mRNA expression of PDL1 in gastric cancer and normal tissues. (E) Relationship between mRNA expression of ANXA2 and PDL1; (F) Expression of PDL1 mRNA in gastric cancer and normal tissues.
Figure 3. miRNAs and TF of ANXA2
(A) Predicted Wayne diagram results of miRNAs; (B) Predicted miRNA expression in gastric cancer and normal tissues and the correlation with ANXA2 expression; (C) Predicted Wayne diagram results of transcription factors; (D) Predicted transcription factor expression in gastric cancer and normal tissues and the correlation with ANXA2 expression; (E) mRNA of SMAD3 expression in relation to the prognosis of gastric cancer patients; (F) Overall relationship among various factors.
Figure 5. Cell proliferation and Flowcytometric results of different cell lines
(A) Cell proliferation in the HGC27 cell line; (B) Cell proliferation in the MKN45 cell line; (C) Flow cytometric analysis of cell cycle in HGC27 NC cells; (D) Flow cytometric analysis of cell cycle in ANXA2-HGC27 cells; (E) Flow cytometric analysis of cell cycle in HGC27 cell line; (F) Flow cytometric analysis of cell cycle in MKN45 NC cells; (G) Plot of flow cytometric analysis of cell cycle of ANXA2-MKN45 cells; (H) Proportion of various cell cycles in MKN45 cells. *P < 0.05, ****P < 0.0001.
Figure 6. Metastatic ability of different cell lines
(A) Images of scratch experiments (100×) for different cell lines; (B) Representative images showing the results from Transwell experiments for various cell lines as specified; (C-D) Bar graphs of statistical results from 24-hour scratch experiments; (E-F) Bar graphs of statistical results from Transwell experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure 7. In vivo results of different cell lines
(A) Photographs of HGC27 cell-implanted mice and the control littermate; (B) Photographs showing the relative tumor sizes between HGC27 cell-implanted mice and the control counterparts; (C) Statistical results of tumor weight between the two groups; (D) Time-dependent changes of tumor volume between the two groups; (E) Relative protein levels in tumor tissues; (F) Luminescence images showing abdominal implantation in mice; (G) Statistical data of luminescence intensity on abdominal implantation tumors. *P < 0.05, ***P < 0.001.
Table 1. ShRNA sequence of NC and ANXA2
ShRNA Strand ShNC Top GATCCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGAATTTTT ShNC Bottom AATTGAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGAATTACGTGACACGTTCGGAGAA ShANXA2 Top CCGGCGGGATGCTTTGAACATTGAATTCAAGAGATTCAATGTTCAAAGCATCCCGTTTTTTG ShANXA2 Bottom AATTCAAAAAACGGGATGCTTTGAACATTGAATCTCTTGAATTCAATGTTCAAAGCATCCCG -
[1] Hajjar K A, Krishnan S. Annexin Ⅱ: a mediator of the plasmin/ plasminogen activator system. Trends Cardiovasc Med, 1999; 9(5): 128-138. doi: 10.1016/S1050-1738(99)00020-1 [2] Morel E, Gruenberg J. Annexin A2 binding to endosomes and functions in endosomal transport are regulated by tyrosine 23 phosphorylation. J Biol Chem, 2009; 284(3): 1604-1611. doi: 10.1074/jbc.M806499200 [3] Deora A B, Kreitzer G, Jacovina A T, et al. An annexin 2 phosphorylation switch mediates p11-dependent translocation of annexin 2 to the cell surface. J Biol Chem, 2004; 279(42): 43411-43418. doi: 10.1074/jbc.M408078200 [4] de Graauw M, Tijdens I, Smeets M B, et al. Annexin A2 phosphorylation mediates cell scattering and branching morphogenesis via cofilin Activation. Mol Cell Biol, 2008; 28(3): 1029-1040. doi: 10.1128/MCB.01247-07 [5] Shiozawa Y, Havens A M, Jung Y, et al. Annexin Ⅱ/annexin Ⅱ receptor axis regulates adhesion, migration, homing, and growth of prostate cancer. J Cell Biochem, 2008; 105(2): 370-380. doi: 10.1002/jcb.21835 [6] Vishwanatha J K, Jindal H K, Davis R G. The role of primer recognition proteins in DNA replication: association with nuclear matrix in HeLa cells. J Cell Sci, 1992; 101 (Pt 1): 25-34. doi: 10.1242/jcs.101.1.25 [7] Kazami T, Nie H, Satoh M, et al. Nuclear accumulation of annexin A2 contributes to chromosomal instability by coilin-mediated centromere damage. Oncogene, 2015; 34(32): 417741-417789. doi: 10.1038/onc.2014.345 [8] Castaldo S A, Ajime T, Serrão G, et al. Annexin A2 regulates AKT upon H2O2-dependent signaling activation in cancer cells. Cancers (Basel), 2019; 11(4): 492. doi: 10.3390/cancers11040492 [9] de Graauw M, Cao L, Winkel L, et al. Annexin A2 depletion delays EGFR endocytic trafficking via cofilin activation and enhances EGFR signaling and metastasis formation. Oncogene, 2014; 33(20): 2610-2619. doi: 10.1038/onc.2013.219 [10] Sarkar S, Swiercz R, Kantara C, et al. Annexin A2 mediates up-regulation of NF-κB, β-catenin, and stem cell in response to progastrin in mice and HEK-293 cells. Gastroenterology, 2011; 140(2): 583-595. e4. doi: 10.1053/j.gastro.2010.08.054 [11] Feng X, Liu H, Zhang Z, et al. Annexin A2 contributes to cisplatin resistance by activation of JNK-p53 pathway in non-small cell lungcancer cells. J Exp Clin Cancer Res, 2017; 36(1): 123. doi: 10.1186/s13046-017-0594-1 [12] Zhang HJ, Yao D F, Yao M, et al. Annexin A2 silencing inhibits invasion, migration, and tumorigenic potential of hepatoma cells. World J Gastroenterol, 2013; 19(24): 3792-3801. doi: 10.3748/wjg.v19.i24.3792 [13] Wang Y X, Lv H, Li Z X, et al. Effect of shRNA mediated downregulation of Annexin A2 on biological behavior of human lung adencarcinoma cells A549. Pathol Oncol Res, 2012; 18(2): 183-190. doi: 10.1007/s12253-011-9427-2 [14] Sharma M R, Rothman V, Tuszynski G P, et al. Antibody-directed targeting of angiostatin's receptor annexin Ⅱ inhibits Lewis Lung Carcinoma tumor growth via blocking of plasminogen activation: possible biochemical mechanism of angiostatin's action. Exp Mol Pathol, 2006; 81(2): 136-145. doi: 10.1016/j.yexmp.2006.03.002 [15] Ling Q, Jacovina A T, Deora A, et al. Annexin Ⅱ regulates fibrin homeostasis and neoangiogenesis in vivo. J Clin Invest, 2004; 113(1): 38-48. doi: 10.1172/JCI19684 [16] Zhai H, Acharya S, Gravanis I, et al. Annexin A2 promotes glioma cell invasion and tumor progression. J Neurosci, 2011; 31(40): 14346-14360. doi: 10.1523/JNEUROSCI.3299-11.2011 [17] Liu Y, Li H, Ban Z, et al. Annexin A2 inhibition suppresses ovarian cancer progression via regulating β-catenin/EMT. Oncol Rep, 2017; 37(6): 3643-3650. doi: 10.3892/or.2017.5578 [18] Rocha M R, Barcellos-de-Souza P, Sousa-Squiavinato A, et al. Annexin A2 overexpression associates with colorectal cancer invasiveness and TGF-ß induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci Rep, 2018; 8(1): 11285. doi: 10.1038/s41598-018-29703-0 [19] Atkinson S J, Gontarczyk A M, Alghamdi A A, et al. The β3-integrin endothelial adhesome regulates microtubule-dependent cell migration. EMBO Rep, 2018; 19(7): e44578. doi: 10.15252/embr.201744578 [20] Wu Y, Cai F, Lu Y, et al. lncRNA RP11-531A24.3 inhibits the migration and proliferation of vascular smooth muscle cells by downregulating ANXA2 expression. Exp Ther Med, 2021; 22(6): 1439. doi: 10.3892/etm.2021.10874 [21] Zhou S, Yi T, Liu R, et al. Proteomics identification of annexin A2 as akey mediator in the metastasis and proangiogenesis of endometrial cells in human adenomyosis. Mol Cell Proteomics, 2012; 11(7): M112.017988. doi: 10.1074/mcp.M112.017988 [22] Chen C Y, Lin Y S, Chen C H, et al. Annexin A2-mediated cancer progression and therapeutic resistance in nasopharyngeal carcinoma. J Biomed Sci, 2018; 25(1): 30. doi: 10.1186/s12929-018-0430-8 [23] Wu B, Zhang F, Yu M, et al. Up-regulation of Anxa2 gene promotes proliferation and invasion of breast cancer MCF-7 cells. Cell Prolif, 2012; 45(3): 189-198. doi: 10.1111/j.1365-2184.2012.00820.x [24] Yao H, Zhang Z, Xiao Z, et al. Identification of metastasis associated proteins in human lung squamous carcinoma using two-dimensional difference gel electrophoresis and laser capture microdissection. Lung Cancer, 2009; 65(1): 41-48. doi: 10.1016/j.lungcan.2008.10.024 [25] Zhang F, Zhang H, Wang Z, et al. P-glycoprotein associates with Anxa2 and promotes invasion in multidrug resistant breast cancer cells. Biochem Pharmacol, 2014; 87(2): 292-302. doi: 10.1016/j.bcp.2013.11.003 [26] Zhang Z D, Li Y, Fan Q, et al. Annexin A2 is implicated in multidrug-resistance in gastric cancer through p38MAPK and AKT pathway. Neoplasma, 2014; 61(6): 627-637. doi: 10.4149/neo_2014_078 [27] Emoto K, Yamada Y, Sawada H, et al. Annexin Ⅱ overexpression correlates with stromal tenascin-C overexpression: a prognostic marker in colorectal carcinoma. Cancer, 2001; 92(6): 1419-1426. doi: 10.1002/1097-0142(20010915)92:6<1419::AID-CNCR1465>3.0.CO;2-J [28] Tas F, Tilgen Yasasever C, Karabulut S, et al. Circulating annexin A2 as a biomarker in gastric cancer patients: correlation with clinical variables. Biomed Pharmacother, 2015; 69: 237-241. doi: 10.1016/j.biopha.2014.12.005 [29] Xie R, Liu J, Yu X, et al. ANXA2 Silencing inhibits proliferation, invasion, and migration in gastric cancer cells. J Oncol, 2019; 2019: 4035460. doi: 10.1155/2019/4035460 [30] Mao L, Yuan W, Cai K, et al. EphA2-YES1-ANXA2 pathway promotes gastric cancer progression and metastasis. Oncogene, 2021; 40(20): 3610-3623. doi: 10.1038/s41388-021-01786-6 [31] Chandrashekar D S, Bashel B, Balasubramanya S, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia, 2017; 19(8): 649-658. doi: 10.1016/j.neo.2017.05.002 [32] Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res, 2021; 23(7): e27633. doi: 10.2196/27633 [33] Asplund A, Edqvist P H, Schwenk J M, et al. Antibodies for profiling the human proteome-the human protein atlas as a resource for cancer research. Proteomics, 2012; 12(13): 2067-2077. doi: 10.1002/pmic.201100504 [34] Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics, 2018; 10(3): 277-288. doi: 10.2217/epi-2017-0118 [35] Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res, 2023; 51(D1): D638-D646. doi: 10.1093/nar/gkac1000 [36] Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun, 2019; 10(1): 1523. doi: 10.1038/s41467-019-09234-6 [37] Li J H, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res, 2014; 42(Database issue): D92-D97. doi: 10.1093/nar/gkt1248 -


 投稿系统
投稿系统


 下载:
下载: