Identification of hub genes and pathways in mouse with cold exposure
doi: 10.1515/fzm-2024-0023
-
Abstract:
Background Cold exposure is linked to numerous diseases, yet the changes in key genes and pathways in mice under cold exposure remain unexplored. Understanding these alterations could offer insights into the mechanisms of cold resistance and contribute valuable ideas for treating cold-related diseases. Methods The dataset GSE148361 was obtained from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were identified using the "limma" package in R software. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on DEGs. The STRING (Search Tool for the Retrieval of Interacting Genes) database was used to construct a protein-protein interaction (PPI) network. Additionally, gene set enrichment analysis (GSEA) was conducted to identify pathways associated with key genes. miRNAs and upstream transcription factors (TFs) were predicted using the miRNet database. Results A total of 208 DEGs were identified, with 137 upregulated and 71 downregulated. In biological processes, DEGs were enriched in nucleotide and purine-containing compound metabolism. For cellular components, DEGs were involved in condensed chromosomes and mitochondrial protein complexes. In molecular functions, proton transmembrane transporter activity was enriched. KEGG pathway analysis showed significant enrichment in biosynthesis of unsaturated fatty acids, fatty acids, and pyruvate metabolism. From the PPI network, 12 hub genes were identified using MCODE. Four hub genes (Col3a1, fi203, Rtp4, Vcan) demonstrated similar trends in a validation set (GSE110420) and were significantly differentially expressed. GSEA analysis indicated that these four genes were enriched in pathways such as ECM-receptor interaction and cytokinecytokine receptor interaction. The hub gene network included 93 miRNAs and one TF. Conclusion This study identified four hub genes as potential diagnostic biomarkers for cold exposure, providing insights for further research on the effects of cold on gene expression and disease. -
Key words:
- cold exposure /
- hub genes /
- pathway /
- adipose tissue
-
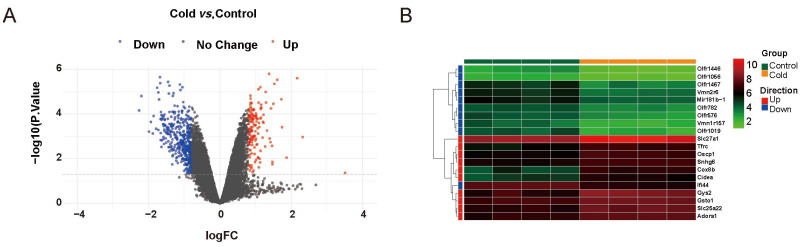
Figure 1. Differential expressed genes identified using LIMMA R package in epididymal white adipose tissue samples between cold exposure and normal conditions with the data set GSE148361
(A) Volcano plot of the differential expressed genes, with blue and red colors representing downregulated and upregulated genes, respectively. (B) Heatmap for the top 10 upregulated genes and top 10 downregulated genes as sorted by adjusted p-value. LIMMA: a library for the analysis of gene expression microarray data, especially the use of linear models for analyzing designed experiments and the assessment of differential expression.
-
[1] Li F X, Liu J J, Xu F, et al. Cold exposure protects against medial arterial calcification development via autophagy. J Nanobiotechnology, 2023; 21(1): 226. doi: 10.1186/s12951-023-01985-1 [2] Christoforou R, Pallubinsky H, Burgholz T M, et al. Influences of indoor air temperatures on empathy and positive affect. Int J Environ Res Public Health, 2024; 21(3): 323. doi: 10.3390/ijerph21030323 [3] Schultze-Rhonhof L, Marzi J, Carvajal Berrio D A, et al. Human tissueresident peritoneal macrophages reveal resistance towards oxidative cell stress induced by non-invasive physical plasma. Front Immunol, 2024; 15: 1357340. doi: 10.3389/fimmu.2024.1357340 [4] Ebrahimi A, Sugiyama A, Ayala-Jacobo L, et al. Integrative analysis of physiology and genomics provides insights into freeze tolerance adaptations of Acacia koa along an elevational cline. Physiol Plant, 2023; 175(6): e14098. doi: 10.1111/ppl.14098 [5] King K E, McCormick J J, Kenny G P. Temperature-dependent relationship of autophagy and apoptotic signaling during cold-water immersion in young and older Males. Adv Biol (Weinh), 2024; 8(3): e2300560. doi: 10.1002/adbi.202300560 [6] Li C, Kiefer M F, Dittrich S, et al. Adipose retinol saturase is regulated by beta-adrenergic signaling and its deletion impairs lipolysis in adipocytes and acute cold tolerance in mice. Mol Metab, 2024; 79: 101855. doi: 10.1016/j.molmet.2023.101855 [7] Neven B. Hereditary systemic autoinflammatory diseases associated with cryopyrin. Rev Prat, 2023; 73(8): 855-862. [8] Puig-Segui M S, Decker C J, Barlit H, et al. Regulation of translation in response to iron deficiency in human cells. Sci Rep, 2024; 14(1): 8451. doi: 10.1038/s41598-024-59003-9 [9] Muratov E, Rosenbaum F P, Fuchs F M, et al. Multifactorial resistance of Bacillus subtilis spores to low-pressure plasma sterilization. Appl Environ Microbiol, 2024; 90(1): e0132923. doi: 10.1128/aem.01329-23 [10] Wallace P J, Gagnon D D, Hartley G L, et al. Effects of skin and mild core cooling on cognitive function in cold air in men. Physiol Rep, 2023; 11(24): e15893. doi: 10.14814/phy2.15893 [11] Du J, He Z, Xu M, et al. Brown adipose tissue rescues bone loss induced by cold exposure. Front Endocrinol (Lausanne), 2021; 12: 778019. [12] Spann R A, Morrison C D, den Hartigh L J. The nuanced metabolic functions of endogenous FGF21 depend on the nature of the stimulus, tissue source, and experimental model. Front Endocrinol (Lausanne). 2021;12: 802541. [13] Zhang Q J, Chen S W, Xu X, et al. The effect of cold exposure on the levels of glucocorticoids, 11-hydroxysteroid dehydrogenase 2, and placental vascularization in a rat model. Eur Rev Med Pharmacol Sci, 2023; 27(24): 11961-11974. [14] Yu G, Wang L G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 2012; 16(5): 284-287. doi: 10.1089/omi.2011.0118 [15] Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: proteinprotein interaction networks, integrated over the tree of life. Nucleic Acids Res, 2015; 43(Database issue): D447-D452. http://broker.edina.ac.uk/782266/1/PMC4383874.pdf [16] Fan Y, Xia J. miRNet-functional analysis and visual exploration of miRNA-target interactions in a network context. Methods Mol Biol, 2018; 1819: 215-233. [17] Li X, Sun X, Kan C, et al. COL1A1: A novel oncogenic gene and therapeutic target in malignancies. Pathol Res Pract, 2022; 236: 154013. doi: 10.1016/j.prp.2022.154013 [18] Denbeigh J M, Hevesi M, Paggi C A, et al. Modernizing storage conditions for fresh osteochondral allografts by optimizing viability at physiologic temperatures and conditions. Cartilage, 2021; 13suppl 1: S280-S292. [19] Choubey D, Walter S, Geng Y, et al. Cytoplasmic localization of the interferon-inducible protein that is encoded by the AIM2 (absent in melanoma) gene from the 200-gene family. FEBS Lett, 2000; 474(1): 3842. [20] Liu M, Goudar CT. Gene expression profiling for mechanistic understanding of cellular aggregation in mammalian cell perfusion cultures. Biotechnol Bioeng, 2013; 110(2): 483-490. doi: 10.1002/bit.24730 [21] Jackson G R, Mowers C C, Sachdev D, et al. Ulnar collateral ligament reconstruction is commonly performed using a palmaris graft and provides favorable patient outcomes with variable return to play and postoperative complication rates: a systematic review. Arthroscopy, 2024; Online ahead of print: S749-S8063. [22] Peebles L A, Blackwood N O, Verma A, et al. Medial ulnar collateral ligament reconstruction with allograft provides excellent clinical outcomes, high rates of return to play, and a low incidence of postoperative complications: a systematic review. Arthroscopy, 2024; Online ahead of print: S749-S8063. [23] Zhou Q, Xu H, Chen W, et al. Controllable blue shift and enhancement emission during the gradually increasing molecular weight of polyacrylamide. Macromol Rapid Commun, 2024; 45(14): e2400073. doi: 10.1002/marc.202400073 [24] Liu F, Chu T, Wang M, et al. Transcriptome analyses provide the first insight into the molecular basis of cold tolerance in Larimichthys polyactis. J Comp Physiol B, 2020; 190(1): 27-34. doi: 10.1007/s00360-019-01247-3 -


 投稿系统
投稿系统


 下载:
下载: