Toll-like receptors 2 polymorphism is associated with psoriasis: A case-control study in the northern Chinese population
doi: 10.1515/fzm-2024-0010
-
Abstract:
Background Psoriasis is a disease caused by genetics and immune system dysfunction, affecting the skin and joints. Toll-like receptors (TLRs) play an important role in triggering the innate immune response and controlling adaptive immunity. The role of TLR2 in the progression of psoriasis is not well understood. Methods A case-control study was conducted on a northern Chinese Han population, consisting of psoriasis patients and healthy control subjects. Genotyping was performed using the tetra-primer amplification refractory mutation system-polymerase chain reaction (ARMS-PCR), and allele and genotype frequencies of four SNPs in TLR2 were analyzed in 270 psoriasis patients and 246 healthy controls. Results Four TLR2 SNPs (rs11938228, rs4696480, rs3804099, rs5743699) were genotyped and found to be in linkage disequilibrium. The genotype distributions of rs11938228 and rs4696480 in two groups were in Hardy-Weinberg equilibrium and statistically significant except for the overdominance model. The haplotypes ATTC and ATCC were found to be protective against psoriasis. Conclusion Our study found a correlation between TLR2 genetic variations and the likelihood of psoriasis in northern China. -
Key words:
- Toll-like receptors 2 /
- psoriasis /
- polymorphism /
- susceptibility
-
Table 1. Sequence of primers
Primer position Primer sequence (5'-3') s11938228 Forward primer GGGGCAAGAATAACCAAGACACA Reverse primer GACATGCCCATATAGGGGTAGATCTTG rs4696480 Forward primer GGTTCTGGAGTCTGGGAAGTCCA Reverse primer ACTGCGAGCTGAGAGGTGGAAC rs3804099 Forward primer TGCAAATCCTGAGAGTGGGAAAT Reverse primer TCCAAACATTCCACGGAACTTG rs5743699 Forward primer AGGATGCCTGGCCCTCTCTACA Reverse primer TGTCTTGGGAATGCAGCCTGTT Table 2. iMLDR probe sequences
SNP allele Primer sequence (5'-3') LDR product s11938228_modify TCTATCCCTATACCAGCAACACACTATATTAAATTTTTTTTTTT 63.21 s11938228_C TTCCGCGTTCGGACTGATATTCAGTGCTATTCTGATCCAATGTTCATGTG 67.77 s11938228_A TACGGTTATTCGGGCTCCTGTTCAGTGCTATTCTGATCCAATGTTCATGTT 66.56 rs4696480_modify GAGGGTCATCTGGCTACATTATAACATGTTTTTTTTTT 63.95 rs4696480_A TCTCTCGGGTCAATTCGTCCTTCAAGATTGAAGGGCTGCATCTCGA 68.65 rs4696480_T TGTTCGTGGGCCGGATTAGTCAAGATTGAAGGGCTGCATCTCGT 67.70 rs3804099_modify GTAAGTCATCTGATCCTTCATATGAAGCATTTTTTTTT 63.00 rs3804099_C TTCCGCGTTCGGACTGATATGAGCCAAAAAGTTTGAAGTCAATTCAGTAC 66.86 rs3804099_T TACGGTTATTCGGGCTCCTGTGAGCCAAAAAGTTTGAAGTCAATTCAGCAT 66.40 rs5743699_modify TGAGCAAAGTCTCTCCGGTTTTTTTTTTTTTT 63.53 rs5743699_C TCTCTCGGGTCAATTCGTCCTTTCTTACTGATATCAATGTTAGTCAAGTTTTTCACAG 64.81 rs5743699_T TGTTCGTGGGCCGGATTAGT TCTTACTGATATCAATGTTAGTCAAGTTTTTCACAA 65.02 Table 3. Demographic characteristics of the studied population
Characteristic Cases, (N = 270) Controls, (N = 247) P Gender, N (%) Male 166 (61.5) 142 (57.5) 0.3706 Female 104 (38.5) 105 (42.5) Age, years 38.17 ± 9.90 37.94 ± 9.70 0.7890 PASI, N (%) ≤ 10 223 (82.6) - - > 10 47 (17.4) - - Age at onset, N (%) ≤ 40 years 219 (81.1) - - > 40 years 51 (18.9) - - Family history, N (%) Yes 102 (37.8) - - No 168 (62.2) - - Data were presented as N(%). Table 4. Allele and genotype frequencies of TLR2 SNPs in the case and control groups
SNP Model Controls Cases OR (95% CI) P P* OR (95% CI) P# rs11938228 Codominant C/C 56 (22.7) 91 (33.7) 1.00 0.00180 0.0072 1.00 0.0016 C/A 124 (50.2) 135 (50.0) 0.67 (0.44-1.01) 0.66 (0.43-1.00) A/A 67 (27.1) 44 (16.3) 0.40 (0.24-0.67) 0.40 (0.24-0.66) C 236 (47.8) 317 (58.7) 1.00 0.00054 0.0022 A 258 (52.2) 223 (41.3) 0.64 (0.50-0.82) Dominant C/C 56 (22.7) 91 (33.7) 1.00 0.00530 0.0212 1.00 0.0041 C/A-A/A 191 (77.3) 179 (66.3) 0.58 (0.39-0.85) 0.57 (0.38-0.84) Recessive C/C-C/A 180 (72.9) 226 (83.7) 1.00 0.00270 0.0108 1.00 0.0027 A/A 67 (27.1) 44 (16.3) 0.52 (0.34-0.80) 0.52 (0.34-0.80) Overdominant C/C-A/A 123 (49.8) 135 (50.0) 1.00 0.96000 1.00 1.00 0.9100 C/A 124 (50.2) 135 (50.0) 0.99 (0.70-1.40) 0.98 (0.69-1.39) rs4696480 Codominant A/A 57 (23.1) 87 (32.2) 1.00 0.01300 0.052 1.00 0.0120 A/T 125 (50.6) 136 (50.4) 0.71 (0.47-1.08) 0.70 (0.46-1.06) T/T 65 (26.3) 47 (17.4) 0.47 (0.29-0.78) 0.47 (0.28-0.78) A 239 (48.4) 310 (57.4) 1.00 0.00400 0.016 T 255 (51.6) 230 (42.6) 0.70(0.54-0.89) Dominant A/A 57 (23.1) 87 (32.2) 1.00 0.02000 0.08 1.00 0.0170 A/T-T/T 190 (76.9) 183 (67.8) 0.63 (0.43-0.93) 0.62 (0.42-0.92) Recessive A/A-A/T 182 (73.7) 223 (82.6) 1.00 0.01400 0.056 1.00 0.0150 T/T 65 (26.3) 47 (17.4) 0.59 (0.39-0.90) 0.59 (0.39-0.91) Overdominant A/A-T/T 122 (49.4) 134 (49.6) 1.00 0.96000 1.00 1.00 0.9000 A/T 125 (50.6) 136 (50.4) 0.99 (0.70-1.40) 0.98 (0.69-1.38) rs3804099 Codominant T/T 123 (49.8) 121 (44.8) 1.00 0.52000 1.00 1.00 0.5300 C/T 106 (42.9) 127 (47.0) 1.22 (0.85-1.75) 1.21 (0.85-1.74) C/C 18 (7.3) 22 (8.2) 1.24 (0.63-2.43) 1.26 (0.64-2.46) T 352 (71.3) 369 (68.3) 1.00 0.30700 1.00 C 142 (28.7) 171 (31.7) 1.15 (0.88-1.50) Dominant T/T 123 (49.8) 121 (44.8) 1.00 0.26000 1.00 1.00 0.2600 C/T-C/C 124 (50.2) 149 (55.2) 1.22 (0.86-1.73) 1.22 (0.86-1.72) Recessive T/T-C/T 229 (92.7) 248 (91.8) 1.00 0.71000 1.00 1.00 0.6800 C/C 18 (7.3) 22 (8.2) 1.13 (0.59-2.16) 1.15 (0.60-2.19) Overdominant T/T-C/C 141 (57.1) 143 (53.0) 1.00 0.35000 1.00 1.00 0.3600 rs5743699 C/T 106 (42.9) 127 (47.0) 1.18 (0.83-1.67) 1.18 (0.83-1.67) C/C 245 (99.2) 268 (99.3) 1.00 0.93000 1.00 1.00 0.8600 C/T 2 (0.8) 2 (0.7) 0.91 (0.13-6.54) 0.84 (0.12-6.05) C 492 (99.6) 538 (99.6) 1.00 0.93000 1.00 T 2 (0.4) 2 (0.4) 0.91 (0.13-6.54) Data were presented as N(%). P: model-based statistical P value, P*: P value adjusted Bonferroni correction, P#: P value adjusted by age and sex; OR: odds ratio; 95% CI: 95% confidence interval. Table 5. Haplotype association with psoriasis
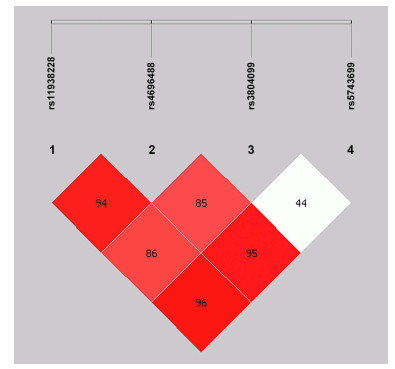
rs11938228 rs4696480 rs3804099 rs5743699 Total Frequency in control Frequency in cases OR (95% CI) P P* OR (95% CI) P# 1 A T T C 0.44 0.48 0.40 0.713 (0.557-0.912) 0.007 0.113 0.705 (0.551-0.902) 0.005 2 C A C C 0.28 0.25 0.30 1.262 (0.960-1.661) 0.095 1.000 1.236 (0.941-1.623) 0.126 3 C A T C 0.24 0.21 0.26 1.299 (0.974-1.732) 0.075 1.000 1.328 (0.994-1.775) 0.054 4 A T C C 0.01 0.02 0.01 0.248 (0.067-0.916) 0.024 0.383 0.225 (0.047-1.068) 0.04 5 C T T C 0.01 0.01 0.01 2.512 (0.623-10.133) 0.180 1.000 2.461 (0.649-9.329) 0.171 6 A A T C 0.01 0.01 0.01 1.289 (0.334-4.975) 0.711 1.000 1.144 (0.305-4.287) 0.84 7 C T C C 0.01 0 0.01 1.836 (0.382-8.831) 0.441 1.000 1.835 (0.334-10.067) 0.477 9 A T T T 0 0 0 0.027 (0.001-0.507) 0.267 1.000 - - P*: P value after Bonferroni correction; P#: the OR and P values were adjusted for age and sex. Table 6. Analysis of linkage disequilibrium based on evaluation of D' in patients
rs11938228 rs4696480 rs3804099 rs5743699 rs11938228 - 0.948 0.869 0.961 rs4696480 - - 0.852 0.956 rs3804099 - - - 0.449 rs5743699 - - - - Table 7. Analysis of linkage disequilibrium based on R2 statistics in patients
rs11938228 rs4696480 rs3804099 rs5743699 rs11938228 - 0.885 0.285 0.004 rs4696480 - - 0.278 0.004 rs3804099 - - - 0 rs5743699 - - - - -
[1] Griffiths C E, Barker J N. Pathogenesis and clinical features of Psoriasis. Lancet, 2007; 370(9583): 263-271. doi: 10.1016/S0140-6736(07)61128-3 [2] Yin X, Low H Q, Wang L, et al. Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat Commun, 2015; 6(1): 6916. doi: 10.1038/ncomms7916 [3] Grän F, Kerstan A, Serfling E, et al. Current developments in the immunology of psoriasis. Yale J Biol Med, 2020; 93(1): 97-110. [4] Conrad C, Gilliet M. Psoriasis: from pathogenesis to targeted therapies. Clin Rev Allergy Immunol, 2018; 54(1): 102-113. doi: 10.1007/s12016-018-8668-1 [5] Miller L S. Toll-like receptors in skin. Adv Dermatol, 2008, 24: 71-87. doi: 10.1016/j.yadr.2008.09.004 [6] Chen J-Q, Szodoray P, Zeher M. Toll-Like receptor pathways in autoimmune diseases. Yale J Biol Med, 2016; 50(1): 1-17. doi: 10.1007/s12016-015-8473-z [7] Ermertcan A T, Öztürk F, Gündüz K. Toll-like receptors and skin: TLRs and skin. Journal of the J Eur Acad Dermatol Venereol, 2011; 25(9): 997-1006. doi: 10.1111/j.1468-3083.2011.04049.x [8] Marks K E, Cho K, Stickling C, et al. Toll-like receptor 2 in autoimmune inflammation. Immune Network, 2021; 21(3): e18. doi: 10.4110/in.2021.21.e18 [9] Garcia-Rodriguez S, Arias-Santiago S, Perandrés-López R, et al. Increased gene expression of Toll-like receptor 4 on peripheral blood mononuclear cells in patients with psoriasis: TLR4 up-regulation in psoriasis PBMC. J Eur Acad Dermatol Venereol, 2013; 27(2): 242-250. doi: 10.1111/j.1468-3083.2011.04372.x [10] Carrasco S, Neves F S, Fonseca M H, et al. Toll-like Receptor (TLR) 2 is upregulated on peripheral blood monocytes of patients with psoriatic arthritis: a role for a gram-positive inflammatory trigger? Clin Exp Rheumatol, 2011; 29(6): 958-962. [11] Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2. A Genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Gene, 2010; 42(11): 985-990. doi: 10.1038/ng.694 [12] Stuart P E, Nair R P, Ellinghaus E, et al. Genome-wide association analysis identifies three psoriasis susceptibility Loci. Nat Gene, 2010, 42(11): 1000-1004. doi: 10.1038/ng.693 [13] Sun L D, Cheng H, Wang Z X, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nate Gene, 2010; 42(11): 1005-1009. doi: 10.1038/ng.690 [14] Collaborative Association Study of Psoriasis (CASP), Genetic Analysis of Psoriasis Consortium, Psoriasis Association Genetics Extension, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate Immunity. Nat Gene, 2012; 44(12): 1341-1348. doi: 10.1038/ng.2467 [15] Shi G, Wang T, Li S, et al. TLR2 and TLR4 polymorphisms in Southern Chinese psoriasis vulgaris patients. J Dermatol Sci, 2016; 83(2): 145-147. doi: 10.1016/j.jdermsci.2016.04.014 [16] Sabah-Özcan S, Gürel G. The polymorphism rs4696480 in the TLR2 gene is associated with psoriasis patients in the Turkish Population. Immunol Let, 2019; 211: 28-32. doi: 10.1016/j.imlet.2019.05.008 [17] Gowin E, Świątek-Kościelna B, Kałużna E, et al. Analysis of TLR2, TLR4, and TLR9 single nucleotide polymorphisms in children with bacterial meningitis and their healthy family Members. Int J Infect Dis, 2017; 60: 23-28. doi: 10.1016/j.ijid.2017.04.024 [18] Zhou B, Liang S, Shang S, et al. Association of TLR2 and TLR9 gene polymorphisms with atopic dermatitis: a systematic review and meta-analysis with trial sequential Analysis. Immunol Med, 2023; 46(1): 32-44. doi: 10.1080/25785826.2022.2132683 [19] Loft N D, Skov L, Iversen L, et al. Associations between functional polymorphisms and response to biological treatment in Danish patients with Psoriasis. Pharmacogenomics J, 2018; 18(3): 494-500. doi: 10.1038/tpj.2017.31 [20] Lorenz E, Mira J P, Cornish K L, et al. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun, 2000; 68(11): 6398-6401. doi: 10.1128/IAI.68.11.6398-6401.2000 [21] Zhou B, Liang S, Shang S, et al. Association of TLR2 and TLR9 gene polymorphisms with atopic dermatitis: a systematic review and meta-analysis with trial sequential Analysis. Immunol Med, 2022: 1-13. [22] Potaczek D P, Przytulska-Szczerbik A, Bazan-Socha S, et al. Interaction between functional polymorphisms in FCER1A and TLR2 and the severity of atopic Dermatitis. Human Immunol, 2020; 81(12): 709-713. doi: 10.1016/j.humimm.2020.08.002 [23] Elloumi N, Tahri S, Fakhfakh R, et al. Role of innate immune receptors TLR4 and TLR2 polymorphisms in systemic lupus erythematosus Susceptibility. Ann Human Gene, 2022; 86(3): 137-144. doi: 10.1111/ahg.12458 [24] Zhang Y, Wang H C, Feng C, et al. Analysis of the association of polymorphisms rs5743708 in TLR2 and rs4986790 in TLR4 with atopic dermatitis risk. Immunol Invest, 2019; 48(2): 169-180. doi: 10.1080/08820139.2018.1508228 [25] Zhang F, Gao X D, Wu W W, et al. Polymorphisms in Toll-like receptors 2, 4 and 5 are associated with legionella pneumophila Infection. Infection, 2013; 41(5): 941-948. doi: 10.1007/s15010-013-0444-9 [26] Shi G, Wang T, Li S, et al. TLR2 and TLR4 polymorphisms in Southern Chinese psoriasis vulgaris patients. J Dermatol Sci, 2016; 83(2): 145-147. doi: 10.1016/j.jdermsci.2016.04.014 -


 投稿系统
投稿系统


 下载:
下载: