Cold exposure alters proteomic profiles of the hypothalamus and pituitary in female rats
doi: 10.2478/fzm-2023-0015
-
Abstract:
Objective Studies have shown that both short-term and long-term cold exposures disturb the biological process. The aim of the present study is to investigate the effects of intermittent cold exposure on proteomic profiles in the hypothalamus and pituitary of female Sprague-Dawley (SD) rats. Materials and methods The rats were exposed to -10℃ in a cabin for 4 h per day, and the treatment lasted for 14 days. The comparative label-free LC-MS/MS analysis was performed to investigate the changes of proteomic profiles in the hypothalamus and pituitary. ELISA analysis was used to validate the expression of differential proteins. Results 22 differential proteins in the hypothalamus and 75 differential proteins in the pituitary were identified by the label-free proteomic analysis. Gene ontology annotation and enrichment analysis indicated that cold exposure disrupted protein phosphorylation, filopodium assembly, intracellular protein transport, peripheral nervous system neuron axonogenesis, spinal cord development, Golgi organization, positive regulation of pseudopodium assembly, and cell-cell adhesion. Three proteins (Cdc42, Ptprs, and Setd7) were down-regulated in the cold exposure group. Conclusion The results indicate that intermittent cold exposure alters the proteomic profiles of hypothalamus and pituitary in female rats. -
Key words:
- cold exposure /
- proteomic profile /
- hypothalamus /
- pituitary
-
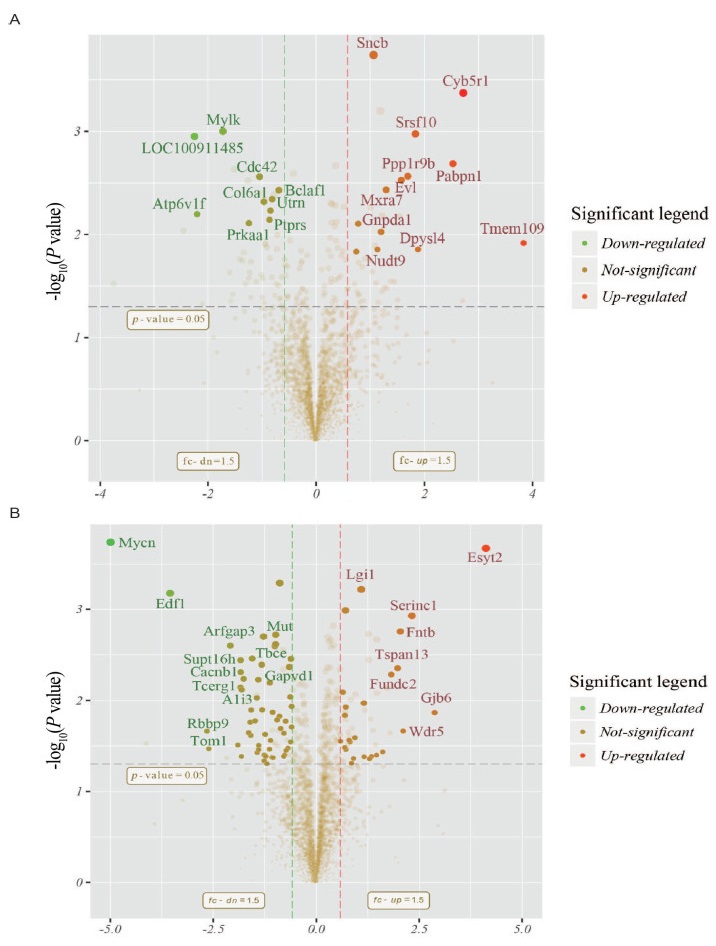
Figure 1. Volcano plot depicting the proteomic alterations in the hypothalamus (A) and pituitary (B)
The red/green colors indicate points-of-interest that display both large magnitude fold-changes (x axis) and high statistical significance (-log10 of P value, y axis). The dashed horizontal line shows the p-value cutoff, and the two vertical dashed lines indicate down-/up-regulated proteins. Transparent points in the significant region means these proteins do not satisfy some other conditions.
Figure 2. Clustering heatmap of the significantly altered proteins in the hypothalamus (A) and pituitary (B) with an FDR < 1% identified by MaxQuant
Red, high expression; blue, low expression. Two main clusters of proteins can be observed, one up-regulated (right) and the other down-regulated (left) in the experimental group. Pearson's distance was used if the sample number equaled to or exceeded 3; otherwiseEuclidean's distance was used. C, control group; CE, cold exposure group.
Figure 3. Enriched GO iterms in the hypothalamus (A) and pituitary (B). Enriched KEGG pathways of the differentially expressed proteins in the pituitary (C)
The number of differentially expressed proteins was shown in the classification of biological process (BP), molecular function (MF) and cellular component (CC). The number of the altered proteins was shown in each KEGG pathway. The P value indicates the degree of enrichment.
Figure 4. STRINGdb protein-protein network enrichment analysis in the hypothalamus (A) and pituitary (B)
Different proteins are represented by nodes of different colors, with straight lines representing protein interactions, dark green and pink for currently known protein interactions, red, blue and green for expected interactions, and light blue, black and light green for other protein interactions.
Figure 5. ELISA analysis of the altered proteins induced by cold exposure in association with the enriched GO terms and KEGG pathways
Data are presented as mean values ± SD of protein levels. Results were normalized by total protein. *P < 0.05 compared to the control group. C, control group; CE, cold exposure group.
Table 1. Differentially expressed proteins in hypothalamus
Entrez ID Protein name Protein description Fold change P-value 100911485 Mfn2 Mitofusin 2; Mitofusin 2, isoform CRA_a 0.21 0.001 1 116664 Atp6v1f V-type proton ATPase subunit F 0.22 0.006 4 288057 Mylk myosin light chain kinase, smooth muscle 0.30 0.001 0 65248 Prkaa1 5'-AMP-activated protein kinase catalytic subunit alpha-1 0.42 0.007 8 64465 Cdc42 Cell division control protein 42 homolog 0.48 0.002 8 25529 Ptprs protein tyrosine phosphatase, receptor type, S 0.55 0.007 2 25600 Utrn utrophin 0.56 0.005 9 293017 Bclaf1 BCL2-associated transcription factor 1 0.57 0.004 5 54238 Cdkn2c cyclin-dependent kinase 4 inhibitor C 0.62 0.003 7 29483 Slc1a3 Excitatory amino acid transporter 1 1.68 0.014 7 362110 Cdk9 Cyclin-dependent kinase 9 1.72 0.007 9 113893 Sncb Beta-synuclein 2.09 0.000 2 305149 Nudt9 ADP-ribose pyrophosphatase, mitochondrial 2.20 0.014 0 683570 Gnpda1 glucosamine-6-phosphate deaminase 1 2.30 0.009 4 690599 Mxra7 matrix-remodelling associated 7 2.45 0.003 7 79115 Evl ena/VASP-like protein 2.97 0.003 0 84686 Ppp1r9b Neurabin-2 3.23 0.002 7 362630 Srsf10 serine/arginine-rich splicing factor 10 3.56 0.001 1 25417 Dpysl4 Dihydropyrimidinase-related protein 4 3.68 0.014 0 116697 Pabpn1 polyadenylate-binding protein 2 5.76 0.002 1 304805 Cyb5r1 NADH-cytochrome b5 reductase 1 6.58 0.000 4 361732 Tmem109 Transmembrane protein 109 14.26 0.012 1 Table 2. Differentially expressed proteins in pituitary
Entrez ID Protein name Protein description Fold change P-value 298894 Mycn N-myc proto-oncogene protein 0.03 0.000 2 296570 Edf1 Endothelial differentiation-related factor 1 0.09 0.000 7 29459 Rbbp9 Putative hydrolase RBBP9 0.16 0.021 7 361370 Tom1 target of Myb protein 1 0.16 0.033 9 503165 Arfgap3 ADP-ribosylation factor GTPase-activating protein 3 0.23 0.002 5 296284 Kif3b kinesin-like protein KIF3B 0.27 0.030 9 307474 Tcerg1 transcription elongation regulator 1 0.28 0.007 2 305851 Supt16h FACT complex subunit SPT16 0.28 0.003 6 50688 Cacnb1 voltage-dependent L-type calcium channel subunit beta-1 0.28 0.004 9 297568 A1i3 alpha-1-inhibitor 3 precursor 0.29 0.007 6 311880 Gapvd1 GTPase activating protein and VPS9 domains 1 0.29 0.005 8 688905 Mtx3 metaxin 3 0.32 0.022 6 360658 Gga3 ADP-ribosylation factor-binding protein GGA3 0.33 0.017 3 361796 Abhd16a Abhydrolase domain-containing protein 16A 0.33 0.024 2 310652 Tdrkh tudor and KH domain containing 0.33 0.012 7 361255 Tbce Tubulin-specific chaperone E 0.34 0.003 5 25081 Apoa1 Apolipoprotein A-I 0.36 0.016 7 361925 Actl6a actin-like protein 6A 0.37 0.009 4 259221 Osbpl1a oxysterol-binding protein-related protein 1 0.38 0.005 9 302500 Mcts1 Malignant T-cell-amplified sequence 1 0.38 0.034 7 306204 Flnb filamin-B 0.38 0.031 2 140594 Sytl4 synaptotagmin-like protein 4 0.40 0.012 7 289233 Pex19 peroxisomal biogenesis factor 19 isoform 1 0.42 0.046 0 294207 Ppp1r11 Protein phosphatase 1 regulatory subunit 11 0.42 0.039 8 64465 Cdc42 Cell division control protein 42 homolog 0.43 0.041 1 295284 Rbm8a RNA-binding protein 8A 0.44 0.049 6 50592 Gria1 glutamate receptor, ionotropic, AMPA1 (alpha 1) 0.45 0.034 3 54301 Slc14a1 Urea transporter 1 0.46 0.006 4 312709 Mlf2 myeloid leukemia factor 2 0.46 0.027 5 108348048 Vwa5a von Willebrand factor A domain containing 5A 0.47 0.042 3 297498 Crbn Protein cereblon 0.48 0.013 5 288704 RGD1311899 Uncharacterized protein C12orf43 homolog 0.48 0.042 6 103690013 Nhp2 NHP2 ribonucleoprotein homolog 0.50 0.002 5 288064 Kpna1 importin subunit alpha-1 0.50 0.002 4 65165 Tmed2 Transmembrane emp24 domain-containing protein 2 0.50 0.001 9 171439 Bzw2 Basic leucine zipper and W2 domain-containing protein 2 0.52 0.016 4 25403 Cast Calpastatin 0.54 0.014 9 171410 Acsbg1 Long-chain-fatty-acid--CoA ligase ACSBG1 0.54 0.000 5 291709 Slc25a46 Solute carrier family 25 member 46 0.55 0.020 3 140667 ENSRNOG00000018650 AP-3 complex subunit mu-2 0.58 0.022 9 296099 Trp53bp1 transformation related protein 53 binding protein 1 0.59 0.039 7 502749 Agk acylglycerol kinase, mitochondrial 0.60 0.016 9 680991 Tubg2 tubulin gamma-2 chain 0.60 0.035 4 171341 Mgst1 Microsomal glutathione S-transferase 1 0.62 0.033 5 306460 Enpp6 Ectonucleotidepyrophosphatase/phosphodiesterase family member 6 0.63 0.004 3 689954 Setd7 histone-lysine N-methyltransferase SETD7 0.65 0.009 1 100910616 Hmgn5 High mobility group nucleosome-binding domain-containing protein 5 0.65 0.028 5 29637 Hmgcs1 Hydroxymethylglutaryl-CoA synthase, cytoplasmic 0.65 0.003 5 293864 Ubl4a Ubiquitin-like protein 4A 0.66 0.019 6 50872 Hpcal4 Hippocalcin-like protein 4 0.66 0.011 6 24688 Prph peripherin 1.50 0.028 0 26197 ATP6 ATP synthase F0 subunit 6 (mitochondrion) 1.56 0.008 1 293863 Lage3 L antigen family member 3 1.62 0.032 9 295586 Clcn6 chloride transport protein 6 1.62 0.014 6 24587 Nefh neurofilament heavy polypeptide 1.64 0.001 0 113960 Cdc42bpb Serine/threonine-protein kinase MRCK beta 1.64 0.011 8 303653 Cdc42ep4 CDC42 effector protein (Rho GTPase binding) 4 1.65 0.034 6 289934 Oxsm 3-oxoacyl-ACP synthase 1.74 0.027 3 298757 Atl2 ADP-ribosylation factor-like 6 interacting protein 2 1.77 0.028 5 290651 Isyna1 Inositol-3-phosphate synthase 1 1.81 0.048 8 100361269 Snrpa1 small nuclear ribonucleoprotein polypeptide A' 1.88 0.043 4 311844 Ccbl1 kynurenine--oxoglutarate transaminase 1, mitochondrial 1.92 0.025 6 252892 Lgi1 Leucine-rich glioma-inactivated protein 1 2.13 0.000 6 81717 Ctbp2 C-terminal-binding protein 2 2.23 0.010 7 363465 Pir Pirin 2.23 0.041 6 108350501 Rps29 40S ribosomal protein S29 2.45 0.043 7 66028 Arl6ip5 PRA1 family protein 3 2.53 0.041 3 300968 Uba5 Ubiquitin-like modifier-activating enzyme 5 2.76 0.039 9 25624 Vamp1 Vesicle-associated membrane protein 1 3.06 0.036 8 361288 Fundc2 FUN14 domain-containing protein 2 3.54 0.005 2 366602 Tspan13 Tetraspanin-13 3.94 0.004 4 64511 Fntb Protein farnesyltransferase subunit beta 4.13 0.001 8 362093 Wdr5 WD repeat-containing protein 5 4.32 0.021 6 294421 Serinc1 Serine incorporator 1 5.00 0.001 2 84403 Gjb6 gap junction beta-6 protein 7.34 0.013 6 -
[1] Wang X, Su M, Qiu Y, et al. Metabolic regulatory network alterations in response to acute cold stress and ginsenoside intervention. J Proteome Res, 2007; 6(9): 3449-3455. doi: 10.1021/pr070051w [2] Manfredi L H, Zanon N M, Garófalo M A, et al. Effect of short-term cold exposure on skeletal muscle protein breakdown in rats. J Appl Physiol, 2013; 115(10): 1496-1505. doi: 10.1152/japplphysiol.00474.2013 [3] Yüksel Ş, Asma D. Effects of extended cold exposure on antioxidant defense system of rat hypothalamic–pituitary–adrenal axis. J Therm Biol, 2006; 31(4): 313-317. doi: 10.1016/j.jtherbio.2005.12.007 [4] Mulligan J D, Gonzalez A A, Stewart A M, et al. Upregulation of AMPK during cold exposure occurs via distinct mechanisms in brown and white adipose tissue of the mouse. J Physiol, 2007; 580(2): 677-684. doi: 10.1113/jphysiol.2007.128652 [5] Nomura T, Kawano F, Kang M S, et al. Effects of long-term cold exposure on contractile muscles of rats. Jpn J Physiol, 2002; 52(1): 85-93. doi: 10.2170/jjphysiol.52.85 [6] Park J J, Lee H, Shin M, et al. Short-term cold exposure may cause a local decrease of neuropeptide Y in the rat hypothalamus. Mol cells, 2007; 23(1): 88-93. [7] Labbé S M, Caron A, Lanfray D, et al. Hypothalamic control of brown adipose tissue thermogenesis. Front Syst Neurosci, 2015; 9: 150. [8] van der Lans A A, Hoeks J, Brans B, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J Clin Invest, 2013; 123(8): 3395-3403. doi: 10.1172/JCI68993 [9] Lee P, Linderman J D, Smith S, et al. Irisin and FGF21 are cold-induced endocrine activators of brown fat function in humans. Cell Metab, 2014; 19(2): 302-309. doi: 10.1016/j.cmet.2013.12.017 [10] Rondeel J M, de Greef W J, Hop W C, et al. Effect of cold exposure on the hypothalamic release of thyrotropin-releasing hormone and catecholamines. Neuroendocrinology, 1991; 54(5): 477-481. doi: 10.1159/000125940 [11] Brissoni B, Agostini L, Kropf M, et al. Intracellular Trafficking of Interleukin-1 Receptor I Requires Tollip. Currt Biol, 2006; 16(22): 2265-2270. doi: 10.1016/j.cub.2006.09.062 [12] Zhao L J, Subramanian T, Zhou Y, et al. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J Biol Chem, 2006; 281(7): 4183-4189. doi: 10.1074/jbc.M509051200 [13] Cao L, Ren Y, Guo X, et al. Downregulation of SETD7 promotes migration and invasion of lung cancer cells via JAK2/STAT3 pathway. Int J Mol Med, 2020; 45(5): 1616-1626. [14] Ramchandran R, Mehta D, Vogel S M, et al. Critical role of Cdc42 in mediating endothelial barrier protection in vivo. Am J Physiol Lung Cell Mol Physiol, 2008; 295(2): L363-369. doi: 10.1152/ajplung.90241.2008 [15] Chen Y, Lin J, Chen J, et al. Mfn2 is involved in intervertebral disc degeneration through autophagy modulation. Osteoarthritis Cartilage, 2020; 28(3): 363-374. doi: 10.1016/j.joca.2019.12.009 [16] Davis T B, Yang M, Schell M J, et al. PTPRS regulates colorectal cancer RAS pathway activity by inactivating Erk and preventing its nuclear translocation. Sci Reps, 2018; 8(1): 9296. doi: 10.1038/s41598-018-27584-x [17] Heasman S J, Ridley A J. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol, 2008; 9(9): 690-701. doi: 10.1038/nrm2476 [18] Castellani J W, Young A J. Human physiological responses to cold exposure: Acute responses and acclimatization to prolonged exposure. Auton Neurosci, 2016; 196: 63-74. doi: 10.1016/j.autneu.2016.02.009 [19] Lei M, Zhang D, Sun Y, et al. Web-based transcriptome analysis determines a sixteen-gene signature and associated drugs on hearing loss patients: A bioinformatics approach. J Clin Lab Anal, 2021; 35(12): e24065. [20] Nobes C D, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell, 1995; 81(1): 53-62. doi: 10.1016/0092-8674(95)90370-4 [21] Dai M X, Zheng X H, Yu J, et al. The Impact of intermittent and repetitive cold stress exposure on endoplasmic reticulum stress and instability of atherosclerotic plaques. Cell Physiol Biochem, 2014; 34(2): 393-404. doi: 10.1159/000363008 [22] He S, Owen D R, Jelinsky S A, et al. Lysine methyltransferase SETD7 (SET7/9) Regulates ROS Signaling through mitochondria and NFE2L2/ARE pathway. Sci Rep, 2015; 5(1): 14368. doi: 10.1038/srep14368 [23] Chouchani E T, Kazak L, Jedrychowski M P, et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature, 2016; 532(7597): 112-116. doi: 10.1038/nature17399 -


 投稿系统
投稿系统


 下载:
下载: