Prospects of DNA microarray application in management of chronic obstructive pulmonary disease: A systematic review
doi: 10.2478/fzm-2023-0002
-
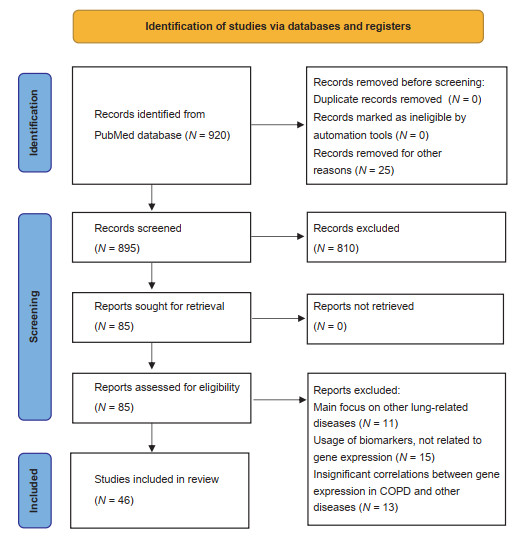
Abstract: Chronic obstructive pulmonary disease (COPD) is incurable chronic disease which kills 3.3 million each year worldwide. Number of global cases of COPD is steadily rising alongside with life expectancy, disproportionally hitting middle-income countries like Russia and China, in such conditions, new approaches to the COPD management are desperately needed. DNA microarray technology is a powerful genomic tool that has the potential to uncover underlying COPD biological alteration and brings up revolutionized treatment option to clinicians. We executed systematic review studies of studies published in last 10 years regarding DNA microarray application in COPD management, with complacence to PRISMA criteria and using PubMed and Medline data bases as data source. Out of 920 identified papers, 39 were included in the final analysis. We concluded that Genome-wide expression profiling using DNA microarray technology has great potential in enhancing COPD management. Current studied proofed this method is reliable and possesses many potential applications such as individual at risk of COPD development recognition, early diagnosis of disease, COPD phenotype identification, exacerbation prediction, personalized treatment optioning and prospect of oncogenesis evaluation in patients with COPD. Despite all the proofed benefits of this technology, researchers are still in the early stage of exploring it's potential. Therefore, large clinical trials are still needed to set up standard for DNA microarray techniques usage implementation in COPD management guidelines, subsequently giving opportunity to clinicians for controlling or even eliminating COPD entirely.
-
Key words:
- chronic obstructive pulmonary disease /
- biomarker /
- expression profiling /
- DNA microarray
-
Table 1. Summary of identified potential genes for COPD diagnosis
Authors Publication year DNA microarray platform Sampling source Significant genes detected Bhattacharya et al.[17] 2011 Affymetrix Peripheral blood, lung tissue RP9, NAPE-PLD, ARID2, STX17, FOXP1, SESN1 Bahr et al.[18] 2013 Affymetrix Peripheral blood, GSE42057 ASAH1, FOXP1, TLR8, VNN2 Wei et al.[19] 2015 Affymetrix GSE29133 HMGCS2, FABP6, TAP1, HLA-A, HLA-DOB, HLA-F Boudewijn et al.[20] 2017 Affymetrix Nasal epithelial brushings NPHP1, CFAP206, CCDC113, MUC1, CREB3L1, DSP Yao et al.[21] 2019 Aglient Peripheral blood HBEGF, DIO2, CLCN3 Rogers et al.[22] 2019 Affymetrix, Aglient Peripheral blood DUSP7, GPR15, PLD1, RPS4Y1, FCGR1B, TCF7 Huang et al.[23] 2019 Affymetrix Lung tissue CX3CR1, PPBP, PTGS2, FPR1, FPR2, VCAM1, S100A12, ARG1, EGR1, CD163, FGG, ORM1, S100A8, S100A9 Yu et al.[24] 2021 Affymetrix Lung tissue MTHFD2, KANK3, GFPT2, PHLDA1, HS3ST2, FGG, RPS4Y1 Winter et al.[25] 2021 Affymetrix Sputum, peripheral blood TPSB2, CPA3, KIT, GATA2, SOCS2, ENO2, GPR56, HDC COPD, chronic obstructive pulmonary disease. -
[1] World Health Organization. The top 10 causes of death. 2020. Available at: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death. Accessed on 03 March, 2022. [2] Adeloye D, Song P, Zhu Y, et al. Global, regional, and national prevalence of, and risk factors for, Chronic obstructive pulmonary disease (COPD) in 2019: a systematic review and modelling analysis. Lancet Respir Med, 2022; 10(5): 447-458. doi: 10.1016/S2213-2600(21)00511-7 [3] Szalontai K, Gémes N, Furák J, et al. Chronic obstructive pulmonary disease: epidemiology, biomarkers, and paving the way to lung cancer. J Clin Med, 2021; 10(13): 2889. doi: 10.3390/jcm10132889 [4] Quan Z, Yan G, Wang Z, et al. Current status and preventive strategies of chronic obstructive pulmonary disease in China: a literature review. J Thorac Dis, 2021; 13(6): 3865-3877. doi: 10.21037/jtd-20-2051 [5] Cook S, Eggen A E, Hopstock L A, et al. Chronic obstructive pulmonary disease (COPD) in population studies in Russia and Norway: comparison of prevalence, awareness and management. Int J Chron Obstruct Pulmon Dis, 2021; 16: 1353-1368. doi: 10.2147/COPD.S292472 [6] Fettes L, Bone A E, Etkind S N, et al. Disability in basic activities of daily living is associated with symptom burden in older people with advanced cancer or chronic obstructive pulmonary disease: a secondary data analysis. J Pain Symptom Manage, 2021; 61(6): 1205-1214. doi: 10.1016/j.jpainsymman.2020.10.012 [7] Gupta N, Malhotra N, Ish P. GOLD 2021 guidelines for COPD - what's new and why. Adv Respir Med, 2021; 89(3): 344-346. doi: 10.5603/ARM.a2021.0015 [8] Taub F E, DeLeo J M, Thompson E B. Sequential comparative hybridizations analyzed by computerized image processing can identify and quantitate regulated RNAs. DNA, 1983; 2(4): 309-327. doi: 10.1089/dna.1983.2.309 [9] Moher D, Shamseer L, Clarke M, et al. PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev, 2015; 4(1): 1. doi: 10.1186/2046-4053-4-1 [10] Centre for Reviews and Dissemination (UK). Database of abstracts of reviews of effects (DARE): quality-assessed reviews. York (UK): Univeristy of York, 1995. [11] Van Gelder R N, Von Zastrow M E, Yool A, et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A, 1990; 87(5): 1663-1667. doi: 10.1073/pnas.87.5.1663 [12] Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment(MIAME) - toward standards for microarray data. Nat Genet, 2001; 29(4): 365-371. doi: 10.1038/ng1201-365 [13] Naidu C N Suneetha Y. Current knowledge on microarray technology-an overview. Trop J Pharm Res, 2012; 11(1): 153-164. [14] Chang Y, Glass K, Liu Y Y, et al. COPD subtypes identified by network-based clustering of blood gene expression. Genomics, 2016; 107(2-3): 51-58. doi: 10.1016/j.ygeno.2016.01.004 [15] Vestbo J, Anderson W, Coxson H O, et al. Evaluation of COPD longitudinally to identify predictive surrogate end-points (ECLIPSE). Eur Respir J, 2008; 31(4): 869-873. doi: 10.1183/09031936.00111707 [16] Baines K J, Negewo N A, Gibson P G, et al. A Sputum 6 gene expression signature predicts inflammatory phenotypes and future exacerbations of COPD. Int J Chron Obstruct Pulmon Dis, 2020; 15: 1577-1590. doi: 10.2147/COPD.S245519 [17] Bhattacharya S, Tyagi S, Srisuma S, et al. Peripheral blood gene expression profiles in COPD subjects. J Clin Bioinforma, 2011; 1(1): 12. doi: 10.1186/2043-9113-1-12 [18] Bahr T M, Hughes G J, Armstrong M, et al. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol, 2013; 49(2): 316-323. doi: 10.1165/rcmb.2012-0230OC [19] Wei L, Xu D, Qian Y, et al Comprehensive analysis of gene-expression profile in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis, 2015; 10: 1103-1109. [20] Boudewijn I M, Faiz A, Steiling K, et al. Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression. Respir Res, 2017; 18(1): 213. doi: 10.1186/s12931-017-0696-5 [21] Yao Y, Gu Y, Yang M, et al. The Gene expression biomarkers for chronic obstructive pulmonary disease and interstitial lung disease. Front Genet, 2019; 10: 1154. doi: 10.3389/fgene.2019.01154 [22] Rogers L R K, Verlinde M, Mias G I. Gene expression microarray public dataset reanalysis in chronic obstructive pulmonary disease. PLoS One, 2019; 14(11): e0224750. doi: 10.1371/journal.pone.0224750 [23] Huang X, Li Y, Guo X, et al. Identification of differentially expressed genes and signaling pathways in chronic obstructive pulmonary disease via bioinformatic analysis. FEBS Open Bio, 2019; 9(11): 1880-1899. doi: 10.1002/2211-5463.12719 [24] Yu H, Guo W, Liu Y, et al. Immune characteristics analysis and transcriptional regulation prediction based on gene signatures of Chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis, 2021; 16: 3027-3039. doi: 10.2147/COPD.S325328 [25] Winter N A, Gibson P G, McDonald V M, et al. Sputum gene expression reveals dysregulation of mast cells and basophils in eosinophilic COPD. Int J Chron Obstruct Pulmon Dis, 2021; 16: 2165-2179. doi: 10.2147/COPD.S305380 [26] Savarimuthu Francis S M, Larsen J E, Pavey S J, et al. Genes and gene ontologies common to airflow obstruction and emphysema in the lungs of patients with COPD. PLoS One, 2011; 6(3): e17442. doi: 10.1371/journal.pone.0017442 [27] Ning W, Li C J, Kaminski N, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A, 2004; 101(41): 14895-14900. doi: 10.1073/pnas.0401168101 [28] Wang I M, Stepaniants S, Boie Y, , et al. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med, 2008; 177(4): 402-411. doi: 10.1164/rccm.200703-390OC [29] Bhattacharya S, Srisuma S, Demeo D L, et al. Molecular biomarkers for quantitative and discrete COPD phenotypes. Am J Respir Cell Mol Biol, 2009; 40(3): 359-367. doi: 10.1165/rcmb.2008-0114OC [30] Singh D, Agusti A, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: the GOLD science committee report 2019. Eur Respir J, 2019; 53(5): 1900164. doi: 10.1183/13993003.00164-2019 [31] Steiling K, van den Berge M, Hijazi K, et al. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med, 2013; 187(9): 933-942. doi: 10.1164/rccm.201208-1449OC [32] Beane J, Sebastiani P, Liu G, et al. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol, 2007; 8(9): R201. doi: 10.1186/gb-2007-8-9-r201 [33] Poliska S, Csanky E, Szanto A, et al. Chronic obstructive pulmonary disease-specific gene expression signatures of alveolar macrophages as well as peripheral blood monocytes overlap and correlate with lung function. Respiration, 2011; 81: 499-510. doi: 10.1159/000324297 [34] Wu Y, Xu B, He X, et al. Correlation between autophagy levels in peripheral blood mononuclear cells and clinical parameters in patients with chronic obstructive pulmonary disease. Mol Med Rep, 2018; 17(6): 8003-8009. [35] Sun S, Shen Y, Wang J, et al. Identification and Validation of Autophagy-Related Genes in Chronic Obstructive Pulmonary Disease. Int J Chron Obstruct Pulmon Dis, 2021; 16: 67-78. doi: 10.2147/COPD.S288428 [36] Zhang J, Zhu C, Gao H, et al. Identification of biomarkers associated with clinical severity of chronic obstructive pulmonary disease. PeerJ, 2020; 8: e10513. doi: 10.7717/peerj.10513 [37] Morrow J D, Qiu W, Chhabra D, et al. Identifying a gene expression signature of frequent COPD exacerbations in peripheral blood using network methods. BMC Med Genomics, 2015; 8: 1. doi: 10.1186/s12920-014-0072-y [38] Gompertz S, O'Brien C, Bayley DL, et al. Changes in bronchial inflammation during acute exacerbations of chronic bronchitis. Eur Respir J, 2001; 17(6): 1112-1119. doi: 10.1183/09031936.01.99114901 [39] Wu X, Sun X, Chen C, et al. Dynamic gene expressions of peripheral blood mononuclear cells in patients with acute exacerbation of chronic obstructive pulmonary disease: a preliminary study. Crit Care, 2014; 18(6): 508. doi: 10.1186/s13054-014-0508-y [40] Singh D, Fox S M, Tal-Singer R, et al. Altered gene expression in blood and sputum in COPD frequent exacerbators in the ECLIPSE cohort. PLoS One, 2014; 9(9): e107381. doi: 10.1371/journal.pone.0107381 [41] Van Den Berge M, Steiling K, Timens W, et al. Airway gene expression in COPD is dynamic with inhaled corticosteroid treatment and reflects biological pathways associated with disease activity. Thorax, 2014; 69(1): 14-23. doi: 10.1136/thoraxjnl-2012-202878 [42] Lee J, Machin M, Russell K E, et al. Corticosteroid modulation of immunoglobulin expression and B-cell function in COPD. FASEB J, 2016; 30(5): 2014-2026. doi: 10.1096/fj.201500135 [43] Rossios C, To Y, Osoata G, et al. Corticosteroid insensitivity is reversed by formoterol via phosphoinositide-3-kinase inhibition. Br J Pharmacol, 2012; 167(4): 775-786. doi: 10.1111/j.1476-5381.2012.01864.x [44] Pei G, Ma N, Chen F, et al. Screening and identification of hub genes in the corticosteroid resistance network in human airway epithelial cells via microarray analysis. Front Pharmacol, 2021; 12: 672065. doi: 10.3389/fphar.2021.672065 [45] Baines K J, Wright T K, Gibson P G, et al. Azithromycin treatment modifies airway and blood gene expression networks in neutrophilic COPD. ERJ Open Res, 2018; 4(4): 00031-2018. [46] Seo M, Qiu W, Bailey W, et al. Genomics and response to long-term oxygen therapy in chronic obstructive pulmonary disease. J Mol Med (Berl), 2018; 96(12): 1375-1385. doi: 10.1007/s00109-018-1708-8 [47] Young R P, Hopkins R J. Link between COPD and lung cancer. Respir Med, 2010; 104(5): 758-759. doi: 10.1016/j.rmed.2009.11.025 [48] Young R P, Hopkins R J, Christmas T, et al. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur Respir J, 2009; 34(2): 380-386. doi: 10.1183/09031936.00144208 [49] Miao T W, Xiao W, Du L Y, et al. High expression of SPP1 in patients with chronic obstructive pulmonary disease (COPD) is correlated with increased risk of lung cancer. FEBS Open Bio, 2021; 11(4): 1237-1249. doi: 10.1002/2211-5463.13127 [50] Sun X, Shang J, Wu A, et al. Identification of dynamic signatures associated with smoking-related squamous cell lung cancer and chronic obstructive pulmonary disease. J Cell Mol Med, 2020; 24(2): 1614-1625. doi: 10.1111/jcmm.14852 [51] Okudela K, Woo T, Mitsui H, et al. Downregulation of ALDH1A1 expression in non-small cell lung carcinomas--its clinicopathologic and biological significance. Int J Clin Exp Pathol, 2013; 6(1): 1-12. [52] Zhang L, Chen J, Yang H, et al. Multiple microarray analyses identify key genes associated with the development of non-small cell lung cancer from Chronic obstructive pulmonary disease. J Cancer, 2021; 12(4): 996-1010. doi: 10.7150/jca.51264 -


 投稿系统
投稿系统


 下载:
下载: