RNA modification by M6A methylation in cardiovascular diseases: Current trends and future directions
doi: 10.2478/fzm-2022-0023
-
Abstract: N6-methyladenosine (M6A) is the most common modification in eukaryotic RNAs for the regulation of RNA transcription, processing, splicing, degradation, and translation. RNA modification by M6A is dynamically reversible, involving methylated transferase, demethylase, and methylated reading protein. M6A-mediated gene regulation involves cell differentiation, metastasis, apoptosis, and proliferation. Dysregulation of M6A can lead to various diseases. Cardiovascular disease (CVD) seriously endangers human health and brings great social burden. Seeking effective prevention and treatment strategies for CVD is a challenge to both fundamentalists and clinicians. Substantial evidence has suggested the key role of M6A modification in the development of CVDs. This review summarizes the mechanism of M6A RNA modification and the latest research progress in respect with its role in CVDs, including atherosclerosis, coronary artery disease, myocardial infarction and cardiac remodeling, myocardial ischemia-reperfusion injury, heart failure, hypertension, and aortic aneurysm, and the potential applications of the findings to CVDs, thereby providing new ideas and approaches for the diagnosis and therapy of CVDs.
-
Key words:
- RNA modification /
- M6A methylation /
- cardiovascular disease /
- epigenetics
-
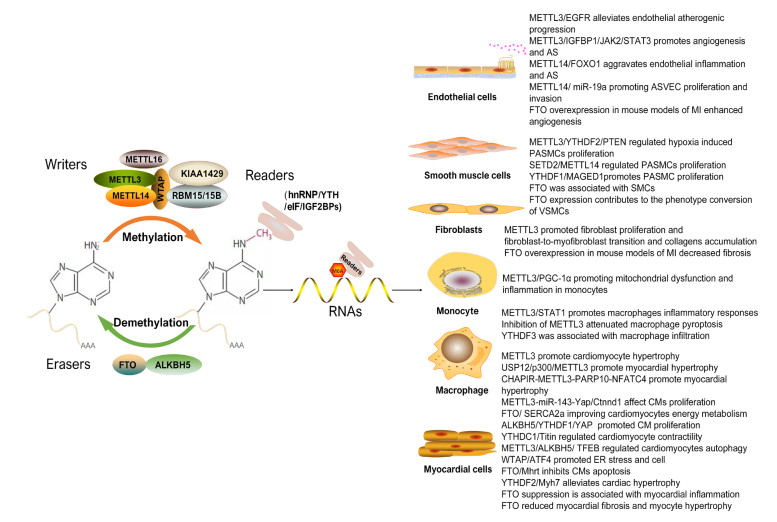
Figure 1. M6A modification and its mechanisms involved in cardiovascular diseases
hnRNP, (nuclear inhomogeneous ribose protein); eIF, eukaryotic initiation factors; IGF2BPs, insulin like growth factor 2 mRNA-binding proteins; EGFR, epidermal growth factor receptor; JAK2, janus kinase 2; STAT3, (activator of transcription 3); FOXO1, forkhead box O1; ASVEC, atherosclerotic vascular endothelial cells; MI, myocardial infarction; PTEN. phosphatase and tensin homolog; SETD2, SET domain-containing 2; PASMCs, pulmonary artery smooth muscle cell; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1-α; STAT1, signal transducer and activator of transcription 1; USP12, ubiquitin specific proteinase 12; Ctnnd1, catenin delta 1; SERCA2a, sarcoplasmic/endoplasmic reticulum calcium ATPase 2a; YAP, yes-associated protein; TFEB, transcription factor EB; ATF4, activating transcription factor 4.
Table 1. The role of M6A regulators in cardiovascular diseases
Cardiovascular disease M6A regulator Related gene Mechanism Atherosclerosis /coronary artery disease METTL3 EGFR METTL3 mitigated endothelial atherogenic progression by M6A-dependent stabilization of EGFR mRNA[62] METTL3 NLRP1/ KLF4 In the AS model, partial ligation of the carotid artery resulted in plaque formation and up-regulation of METTL3. Knockdown of METTL3 prevented the atherogenic process[61] METTL3 STAT1 METTL3 promotes ox-LDL-triggered inflammatory responses in macrophages by interacting with STAT1 protein and mRNA[58] METTL3 IRF-1/ hsa_circ_0029589 Overexpression of IRF-1 suppressed the expression of hsa_circ_0029589, but induced its M6A level along with the expression of METTL3 in macrophages. Overexpression of hsa_circ_0029589 or inhibition of METTL3 significantly increased the expression of hsa_circ_0029589 and attenuated macrophage pyroptosis[59] METTL3 PGC-1 METTL3 modifies PGC-1α mRNA promoting mitochondrial dysfunction and ox-LDL-induced inflammation in monocytes[57] METTL3 JAK2 METTL3 knockdown prevented AS progression by inhibiting JAK2/STAT3 pathway via IGF2BP1[60] METTL14 FOXO1 METTL14 aggravated endothelial inflammation and AS by increasing FOXO1 M6A modification[64] METTL14 miR-19a METTL14 increased the M6A modification of pri-miR-19a and promoted the processing of mature miR-19a, thus promoting the proliferation and invasion of ASVEC[63] FTO Related to neointima formation[56] ALKBH1 MIAT/ HIF1α Silencing of ALKBH1 or HIF1α could rescue the ox-LDL-increased level of MIAT[55] YTHDF1 NLRP1 The METTL3-mediated RNA hypermethylation up-regulated NLRP1 transcript through YTHDF1[61] YTHDF2 KLF4 The METTL3-mediated RNA hypermethylation down-regulated KLF4 transcript through YTHDF2[61] YTHDF2 PGC-1 METTL3 coordinated with YTHDF2 to suppress the expression of PGC-1α, as well as CYCS and NDUFC2 and reduced ATP production and OCR, which subsequently increased the accumulation of cellular and mitochondrial ROS and the levels of proinflammatory cytokines in inflammatory monocytes[57] IGF2BP1 JAK2 METTL3 knockdown prevented AS progression by inhibiting JAK2/STAT3 pathway via IGF2BP1[60] Myocardial infarction/cardiac remodeling METTL3 Inhibition of METTL3 completely eliminated the ability of cardiomyocytes to undergo hypertrophy when stimulated to grow, whereas increased expression of METTL3 promoted cardiomyocyte hypertrophy both in vitro and in vivo. Cardiac-specific METTL3 knockout mice shown morphological and functional signs of HF with aging and stress[72] METTL3 Collagen-associated genes Enforced expression of METTL3 promoted proliferation and fibroblast-to-myofibroblast transition and collagens accumulation. Silencing METTL3 reduced cardiac fibrosis induced by MI via inhibiting the activation of cardiac fibroblasts[71] METTL3 USP12 was partially dependent on the stabilization of p300 to activate METTL3 expression and promoted myocardial hypertrophy[73] METTL3 CHAPIR The piRNA CHAPIR regulated cardiac hypertrophy and cardiac remodelling by controlling METTL3-dependent M6A of Parp10 mRNA[74] METTL3 miR-143-3p METTL3 deficiency resulted in heart regeneration after MI via METTL3-pri-miR-143-(miR-143)-Yap/Ctnnd1 axis[76] ALKBH5 YAP ALKBH5 regulated cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1[77] YTHDF1 YAP ALKBH5 regulated cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1[77] YTHDC1 Titin Cardiac-specific conditional YTHDC1 knockout led to obvious left ventricular chamber enlargement and severe systolic dysfunction. YTHDC1 induces DCM by abnormal splicing of Titin[75] Myocardial ischemia-reperfusion injury METTL3 BAX /PTEN Down-regulated in both young and elderly hearts. BAX and PTEN are target genes of METTL3 under iH/R stress[81] METTL3 miR-25-3p/miR-873-5p METTL3 up-regulated miR-25-3p and miR-873-5p to activate the PI3K/Akt pathway, leading to the suppression of I/R injury[82] METTL3 TFEB Silencing METTL3 enhanced autophagic flux and inhibited apoptosis in H/R-treated cardiomyocyte[84] WTAP ATF4 WTAP promoted myocardial H/R injury by increasing endoplasmic reticulum stress via regulating M6A modification of ATF4 mRNA[83] FTO SERCA2a FTO decreased M6A level of SERCA2a mRNA, thus accelerating SERCA2a expression, maintaining calcium homeostasis and improving the energy metabolism of H/R cardiomyocytes[79] ALKBH5 Ferritin nanocage loaded with ALKBH5 inhibitor improved the cardiac function and reduced the infarct size in AMI[80] Heart failure FTO Cardiomyocyte restricted knockout of FTO impaired mice cardiac function[51] FTO Up-regulated expression in HFpEF patients and HFpEF mice[89] FTO FTO weakened cardiac dysfunction by regulating glucose uptake and glycolysis in mice with pressure overload-induced HF[90] FTO FTO was decreased expression in failing mammalian hearts and hypoxic cardiomyocytes, thereby increasing M6A in RNA and decreasing cardiomyocyte contractile function. FTO overexpression decreased fibrosis and enhanced angiogenesis[91] FTO MHRT FTO overexpression inhibited apoptosis of hypoxia/reoxygenation-treated myocardial cells by regulating M6A modification of MHRT[92] YTHDF2 MYH7 YTHDF2 improved cardiac hypertrophy by regulating MYH7 mRNA decoy[93] Hypertension METTL3 Down-regulated in postnatal HPH[97] METTL3 PTEN YTHDF2 recognized METTL3 mediated M6A modified PTEN mRNA and accelerated the degradation of PTEN, which resulting in over-proliferation of PASMC by activating PI3K/Akt signaling pathway[99] METTL14 Down-regulated in postnatal HPH[97] METTL14 SETD2 SEDT2/METTL14-mediated M6A methylation awakening resulted in hypoxia-induced PAH in mice[100] FTO Decreased expression of FTO in small PA of MCT-PAH rat[97] FTO Down-regulated in postnatal HPH[97] ALKBH5 Down-regulated in postnatal HPH[97] YTHDF1 MAGED1 YTHDF1 promoted PASMC proliferation and PH by improving MAGED1 translation[98] YTHDF1 Increased expression of YTHDF1 in small PA of MCT-PAH rat[97] YTHDF2 PTEN YTHDF2 recognized METTL3 mediated M6A modified PTEN mRNA and accelerated the degradation of PTEN, which resulting in over-proliferation of PASMC by activating PI3K/Akt signaling pathway[99] Aortic aneurysm METTL3 miR-34a METTL3 induced AAA development and progression by modulating M6A-dependent primary miR-34a processing[105] METTL14 Down-regulated expression in human AAA, low METTL14 expression was related to high WBC and CRP expression[104] METTL14 Associated with inflammatory infiltration and neovascularization[103] RBM15B Up-regulated expression in human AAA[104] FTO KLF5 FTO expression promotes phenotype conversion of VSMC[106] FTO Associated with aneurysm smooth muscle cells[103] YTHDF3 YTHDF3 was associated with a greater risk of rupture and a strong association with macrophage infiltration[103] HNRNPC Down-regulated expression in human AAA[104] Other cardiovascular diseases FTO Overexpression FTO can reduce cardiac fibrosis and myocardial cell hypertrophy[107] FTO CD36 FTO knockdown affects the stability of CD36 mRNA and thus reduces the expression of CD36 and inhibits palmitic acid-induced cardiac inflammation[108] FTO IL-6/TNF-α FTO knockdown in rat cardiomyocyte up-regulated M6A RNA methylation and expression of IL-6 and TNF-α [109] EGFR, epidermal growth factor receptor; M6A, N6-methyladenosine; NLRP1, NLR family pyrin domain containing 1; KLF4, kruppel-like factor 4; AS, atherosclerosis; STAT1, signal transducer and activator of transcription 1; PGC-1α, proliferator-activated receptor γ coactivator 1-α; JAK2, janus kinase 2; STAT3, signal transducer and activator of transcription 3; IGF2BPs, insulin like growth factor 2 mRNA-binding proteins; FOXO1, forkhead box O1; ASVEC, atherosclerotic vascular endothelial cells; HIF1α, hypoxia inducible factor 1α; OCR, oxygen consumption rate; ROS, reactive oxygen species; YAP, yes-associated protein; HF, heart failure; MI, myocardial infarction; USP12, ubiquitin specific proteinase 12; PTEN, phosphatase and tensin homolog; TFEB, transcription factor EB; H/R: hypoxia/reoxygenation; BAX: BCL-2-associated X; SERCA2a, sarcoplasmic/endoplasmic reticulum calcium ATPase 2a; AMI, acute myocardial infarction; HFpEF, heart failure with preserved ejection fraction; HPH, hypoxia mediated pulmonary hypertension; SETD2, SET domain-containing 2; MAGED1, melanoma antigen D1; AAA, abdominal aortic aneurysm; KLF5, kruppel-like factor 5; M6A, N6-methyladenosine. -
[1] Vancheri F, Tate A R, Henein M, et al. Time trends in ischaemic heart disease incidence and mortality over three decades (1990-2019) in 20 Western European countries: systematic analysis of the Global Burden of Disease Study 2019. Eur J Prev Cardiol, 2022; 29(2): 396-403. doi: 10.1093/eurjpc/zwab134 [2] Estruch R, Ruilope L M, Cosentino F. The year in cardiovascular medicine 2020: epidemiology and prevention. Eur Heart J, 2021; 42(8): 813-821. doi: 10.1093/eurheartj/ehaa1062 [3] Fares A. Winter cardiovascular diseases phenomenon. N Am J Med Sci, 2013; 5(4): 266-279 doi: 10.4103/1947-2714.110430 [4] Rumana N, Kita Y, Turin T C, et al. Seasonal pattern of incidence and case fatality of acute myocardial infarction in a Japanese population (from the Takashima AMI Registry, 1988 to 2003). Am J Cardiol, 2008; 102(10): 1307-1311. doi: 10.1016/j.amjcard.2008.07.005 [5] Spencer F A, Goldberg R J, Becker R C, et al. Seasonal distribution of acute myocardial infarction in the second nationalregistry of myocardial infarction. J Am Coll Cardiol, 1998; 31(6): 1226-1233. doi: 10.1016/S0735-1097(98)00098-9 [6] Yang J, Yin P, Zhou M, et al. Cardiovascular mortality risk attributable to ambient temperature in China. Heart, 2015; 101(24): 1966-1972. doi: 10.1136/heartjnl-2015-308062 [7] Kysely J, Pokorna L, Kyncl J, et al. Excess cardiovascular mortality associated with cold spells in the Czech Republic. BMC Public Health, 2009; 9: 19. doi: 10.1186/1471-2458-9-19 [8] Zhai G, Qi J, Zhang X, et al. A comparison of the effect of diurnal temperature range and apparent temperature on cardiovascular disease among farmers in Qingyang, Northwest China. Environ Sci Pollut Res Int, 2022; 29(19): 28946-28956. doi: 10.1007/s11356-021-17785-9 [9] Bhaskaran K, Hajat S, Haines A, et al. Effects of ambient temperature on the incidence of myocardial infarction. Heart, 2009; 95(21): 1760-1769. doi: 10.1136/hrt.2009.175000 [10] Li C Y, Wu P J, Chang C J, et al. Weather impact on acute myocardial infarction hospital admissions with a new model for prediction: a nationwide study. Front Cardiovasc Med, 2021; 8: 725419. doi: 10.3389/fcvm.2021.725419 [11] Sanchez-Gloria J L, Carbo R, Buelna-Chontal M, et al. Cold exposure aggravates pulmonary arterial hypertension through increased miR-146a-5p, miR-155-5p and cytokines TNF-alpha, IL-1beta, and IL-6. Life Sci, 2021; 287: 120091. doi: 10.1016/j.lfs.2021.120091 [12] Virani S S, Alonso A, Benjamin E J, et al. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation, 2020; 141(9): e139-e596. [13] Zhang W, Song M, Qu J, et al. Epigenetic modifications in cardiovascular aging and diseases. Circ Res, 2018, 123(7): 773-786. doi: 10.1161/CIRCRESAHA.118.312497 [14] Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell, 2012; 148(6): 1242-1257. doi: 10.1016/j.cell.2012.03.001 [15] Sabatine M S, Giugliano R P, Keech A C, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med, 2017; 376(18): 1713-1722. doi: 10.1056/NEJMoa1615664 [16] Schwartz G G, Steg P G, Szarek M, et al. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med, 2018; 379(22): 2097-2107. doi: 10.1056/NEJMoa1801174 [17] Ridker P M, Devalaraja M, Baeres F M M, et al. IL-6 inhibition with ziltivekimab in patients at high atherosclerotic risk (RESCUE): a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet, 2021; 397(10289): 2060-2069. doi: 10.1016/S0140-6736(21)00520-1 [18] Ridker P M, Everett B M, Thuren T, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med, 2017; 377(12): 1119-1131. doi: 10.1056/NEJMoa1707914 [19] Ridker P M, MacFadyen J G, Thuren T, et al. Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, doubleblind, placebo-controlled trial. Lancet, 2017; 390(10105): 1833-1842. doi: 10.1016/S0140-6736(17)32247-X [20] Li X, Yang Y, Chen S, et al. Epigenetics-based therapeutics for myocardial fibrosis. Life Sci, 2021; 271: 119186. doi: 10.1016/j.lfs.2021.119186 [21] Wang W, Shao F, Yang X, et al. Author correction: METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N(6)-methyladenosine-dependent YTHDF binding. Nat Commun, 2021; 12(1): 4529. doi: 10.1038/s41467-021-24860-9 [22] Shafik A M, Zhang F, Guo Z, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol, 2021, 22(1): 17. doi: 10.1186/s13059-020-02249-z [23] Komal S, Zhang L R, Han S N. Potential regulatory role of epigenetic RNA methylation in cardiovascular diseases. Biomed Pharmacother, 2021; 137: 111376. doi: 10.1016/j.biopha.2021.111376 [24] Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A, 1974; 71(10): 3971-3975. doi: 10.1073/pnas.71.10.3971 [25] Meyer K D, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell, 2012; 149(7): 1635-1646. doi: 10.1016/j.cell.2012.05.003 [26] Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 2012; 485(7397): 201-206. doi: 10.1038/nature11112 [27] Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell, 2019; 74(4): 640-650. doi: 10.1016/j.molcel.2019.04.025 [28] Ma S, Chen C, Ji X, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol, 2019; 12(1): 121. doi: 10.1186/s13045-019-0805-7 [29] He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer, 2019; 18(1): 176. doi: 10.1186/s12943-019-1109-9 [30] Huisman B, Manske G, Carney S, et al. Functional dissection of the m6A RNA modification. Trends Biochem Sci, 2017; 42(2): 85-86. doi: 10.1016/j.tibs.2016.12.004 [31] Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res, 2018; 28(5): 507-517. doi: 10.1038/s41422-018-0034-6 [32] Scholler E, Weichmann F, Treiber T, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA, 2018; 24(4): 499-512. doi: 10.1261/rna.064063.117 [33] Bokar J A, Shambaugh M E, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA, 1997; 3(11): 1233-1247. [34] Pendleton K E, Chen B, Liu K, et al. The U6 snRNA m(6) A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell, 2017; 169(5): 824-835. doi: 10.1016/j.cell.2017.05.003 [35] Frayling T M, Timpson N J, Weedon M N, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science, 2007; 316(5826): 889-894. doi: 10.1126/science.1141634 [36] Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet, 2007; 39(6): 724-726. doi: 10.1038/ng2048 [37] Scuteri A, Sanna S, Chen W M, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet, 2007; 3(7): e115. doi: 10.1371/journal.pgen.0030115 [38] Fischer J, Koch L, Emmerling C, et al. Inactivation of the FTO gene protects from obesity. Nature, 2009; 458(7240): 894-898. doi: 10.1038/nature07848 [39] Church C, Lee S, Bagg E A, et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet, 2009; 5(8): e1000599. doi: 10.1371/journal.pgen.1000599 [40] Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol, 2011; 7(12): 885-887. doi: 10.1038/nchembio.687 [41] Zheng G, Dahl J A, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell, 2013; 49(1): 18-29. doi: 10.1016/j.molcel.2012.10.015 [42] Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci, 2002; 27(10): 495-497. doi: 10.1016/S0968-0004(02)02189-8 [43] Theler D, Dominguez C, Blatter M, et al. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res, 2014; 42(22): 13911-13919. doi: 10.1093/nar/gku1116 [44] Xu C, Wang X, Liu K, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol, 2014; 10(11): 927-929. doi: 10.1038/nchembio.1654 [45] Meyer K D, Patil D P, Zhou J, et al. 5' UTR m(6)A promotes capindependent translation. Cell, 2015; 163(4): 999-1010. doi: 10.1016/j.cell.2015.10.012 [46] Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol, 2018; 20(3): 285-295. doi: 10.1038/s41556-018-0045-z [47] Patil D P, Chen C K, Pickering B F, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature, 2016; 537(7620): 369-373. doi: 10.1038/nature19342 [48] Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell, 2016; 61(4): 507-519. doi: 10.1016/j.molcel.2016.01.012 [49] Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 2014; 505(7481): 117-120. doi: 10.1038/nature12730 [50] Shen L, Song C X, He C, et al. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem, 2014; 83: 585-614. doi: 10.1146/annurev-biochem-060713-035513 [51] Berulava T, Buchholz E, Elerdashvili V, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail, 2020; 22(1): 54-66. doi: 10.1002/ejhf.1672 [52] Kmietczyk V, Riechert E, Kalinski L, et al. m(6)A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci Alliance, 2019; 2(2): e201800233. [53] Fu J, Cui X, Zhang X, et al. The role of m6A ribonucleic acid modification in the occurrence of atherosclerosis. Front Genet, 2021; 12: 733871. doi: 10.3389/fgene.2021.733871 [54] Quiles-Jimenez A, Gregersen I, de Sousa M M L, et al. N6-methyladenosine in RNA of atherosclerotic plaques: An epitranscriptomic signature of human carotid atherosclerosis. Biochem Biophys Res Commun, 2020; 533(4): 631-637. doi: 10.1016/j.bbrc.2020.09.057 [55] Wu L, Pei Y, Zhu Y, et al. Association of N(6)-methyladenine DNA with plaque progression in atherosclerosis via myocardial infarctionassociated transcripts. Cell Death Dis, 2019; 10(12): 909. doi: 10.1038/s41419-019-2152-6 [56] Deng K, Ning X, Ren X, et al. Transcriptome-wide N6-methyladenosine methylation landscape of coronary artery disease. Epigenomics, 2021; 13(10): 793-808. doi: 10.2217/epi-2020-0372 [57] Zhang X, Li X, Jia H, et al. The m(6)A methyltransferase METTL3 modifies PGC-1alpha mRNA promoting mitochondrial dysfunction and oxLDL-induced inflammation in monocytes. J Biol Chem, 2021; 297(3): 101058. doi: 10.1016/j.jbc.2021.101058 [58] Li Z, Xu Q, Huangfu N, et al. Mettl3 promotes oxLDL-mediated inflammation through activating STAT1 signaling. J Clin Lab Anal, 2022; 36(1): e24019. [59] Guo M, Yan R, Ji Q, et al. IFN regulatory Factor-1 induced macrophage pyroptosis by modulating m6A modification of circ_0029589 in patients with acute coronary syndrome. Int Immunopharmacol, 2020; 86: 106800. doi: 10.1016/j.intimp.2020.106800 [60] Dong G, Yu J, han G, et al. N6-Methyladenosine methyltransferase METTL3 promotes angiogenesis and atherosclerosis by upregulating the JAK2/STAT3 pathway via m6A reader IGF2BP1. Front Cell Dev Biol, 2021; 9: 731810. doi: 10.3389/fcell.2021.731810 [61] Chien C S, Li J Y, Chien Y, et al. METTL3-dependent N(6)-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U S A, 2021; 118(7): e2025070118. doi: 10.1073/pnas.2025070118 [62] Li B, Zhang T, Liu M, et al. RNA N(6)-methyladenosine modulates endothelial atherogenic responses to disturbed flow in mice. Elife, 2022; 11: e69906. doi: 10.7554/eLife.69906 [63] Zhang B Y, Han L, Tang Y F, et al. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur Rev Med Pharmacol Sci, 2020; 24(12): 7015-7023. [64] Jian D, Wang Y, Jian L, et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics, 2020; 10(20): 8939-8956. doi: 10.7150/thno.45178 [65] Tang X, Yin R, Shi H, et al. LncRNA ZFAS1 confers inflammatory responses and reduces cholesterol efflux in atherosclerosis through regulating miR-654-3p-ADAM10/RAB22A axis. Int J Cardiol, 2020; 315: 72-80. doi: 10.1016/j.ijcard.2020.03.056 [66] Chen L, Yao H, Hui J Y, et al. Global transcriptomic study of atherosclerosis development in rats. Gene, 2016; 592(1): 43-48. doi: 10.1016/j.gene.2016.07.023 [67] Gong C, Fan Y, Liu J. METTL14 mediated m6A modification to LncRNA ZFAS1/RAB22A: A novel therapeutic target for atherosclerosis. Int J Cardiol, 2021; 328: 177. doi: 10.1016/j.ijcard.2020.12.002 [68] Chang J S, Lin Z X, Liu Y J, et al. Ultra performance liquid chromatography-tandem mass spectrometry assay for the quantification of RNA and DNA methylation. J Pharm Biomed Anal, 2021; 197: 113969. doi: 10.1016/j.jpba.2021.113969 [69] Shi X, Cao Y, Zhang X, et al. Comprehensive analysis of N6-Methyladenosine RNA methylation regulators expression identify distinct molecular subtypes of myocardial infarction. Front Cell Dev Biol, 2021; 9: 756483. doi: 10.3389/fcell.2021.756483 [70] Alarcon C R, Goodarzi H, Lee H, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell, 2015, 162(6): 1299-1308. doi: 10.1016/j.cell.2015.08.011 [71] T. Li, Y. Zhuang, W. Yang, et al. Silencing of METTL3 attenuates cardiac fibrosis induced by myocardial infarction via inhibiting the activation of cardiac fibroblasts. FASEB J, 2021, 35(2): e21162. [72] Dorn L E, Lasman L, Chen J, et al. The N(6)-Methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation, 2019; 139(4): 533-545. doi: 10.1161/CIRCULATIONAHA.118.036146 [73] Lu P, Xu Y, Sheng Z Y, et al. De-ubiquitination of p300 by USP12 Critically Enhances METTL3 Expression and Ang Ⅱ-induced cardiac hypertrophy. Exp Cell Res, 2021; 406(1): 112761. doi: 10.1016/j.yexcr.2021.112761 [74] Gao X Q, Zhang Y H, Liu F, et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N(6)-methyladenosine methylation of Parp10 mRNA. Nat Cell Biol, 2020; 22(11): 1319-1331. doi: 10.1038/s41556-020-0576-y [75] Gao S, Sun H, Chen K, et al. Depletion of m(6) A reader protein YTHDC1 induces dilated cardiomyopathy by abnormal splicing of Titin. J Cell Mol Med, 2021; 25(23): 10879-10891. doi: 10.1111/jcmm.16955 [76] Gong R, Wang X, Li H, et al. Loss of m(6)A methyltransferase METTL3 promotes heart regeneration and repair after myocardial injury. Pharmacol Res, 2021; 174: 105845. doi: 10.1016/j.phrs.2021.105845 [77] Han Z, Wang X, Xu Z, et al. ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics, 2021; 11(6): 3000-3016. doi: 10.7150/thno.47354 [78] Wang K, Li Y, Qiang T, et al. Role of epigenetic regulation in myocardial ischemia/reperfusion injury. Pharmacol Res, 2021; 170: 105743. doi: 10.1016/j.phrs.2021.105743 [79] Deng W, Jin Q, Li L. Protective mechanism of demethylase fat mass and obesity-associated protein in energy metabolism disorder of hypoxia-reoxygenation-induced cardiomyocytes. Exp Physiol, 2021; 106(12): 2423-2433. doi: 10.1113/EP089901 [80] Cheng P, Han H, Chen F, et al. Amelioration of acute myocardial infarction injury through targeted ferritin nanocages loaded with an ALKBH5 inhibitor. Acta Biomater, 2021; 140: 481-491. [81] Su X, Shen Y, Jin Y, et al. Aging-associated differences in epitranscriptomic m6A regulation in response to acute cardiac ischemia/reperfusion injury in female mice. Front Pharmacol, 2021; 12: 654316. doi: 10.3389/fphar.2021.654316 [82] Zhao X, Yang L, Qin L. Methyltransferase-like 3 (METTL3) attenuates cardiomyocyte apoptosis with myocardial ischemiareperfusion (I/R) injury through miR-25-3p and miR-873-5p. Cell Biol Int, 2021: 11706. [83] Wang J, Zhang J, Ma Y, et al. WTAP promotes myocardial ischemia/reperfusion injury by increasing endoplasmic reticulum stress via regulating m(6)A modification of ATF4 mRNA. Aging (Albany NY), 2021; 13(8): 11135-11149. [84] Song H, Feng X, Zhang H, et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy, 2019; 15(8): 1419-1437. doi: 10.1080/15548627.2019.1586246 [85] Ruan Z, Wang S, Yu W, et al. LncRNA MALAT1 aggravates inflammation response through regulating PTGS2 by targeting miR-26b in myocardial ischemia-reperfusion injury. Int J Cardiol, 2019; 288: 122. doi: 10.1016/j.ijcard.2019.04.015 [86] Zhao Z H, Hao W, Meng Q T, et al. Long non-coding RNA MALAT1 functions as a mediator in cardioprotective effects of fentanyl in myocardial ischemia-reperfusion injury. Cell Biol Int, 2017; 41(1): 62-70. doi: 10.1002/cbin.10701 [87] Yang C, Fan Z, Yang J. m(6)A modification of LncRNA MALAT1: A novel therapeutic target for myocardial ischemia-reperfusion injury. Int J Cardiol, 2020; 306: 162. doi: 10.1016/j.ijcard.2019.11.140 [88] Hinger S A, Wei J, Dorn L E, et al. Remodeling of the m(6)A landscape in the heart reveals few conserved post-transcriptional events underlying cardiomyocyte hypertrophy. J Mol Cell Cardiol, 2021; 151: 46-55. doi: 10.1016/j.yjmcc.2020.11.002 [89] Zhang B, Xu Y, Cui X, et al. Alteration of m6A RNA Methylation in Heart Failure With Preserved Ejection Fraction. Front Cardiovasc Med, 2021, 8: 647806. doi: 10.3389/fcvm.2021.647806 [90] Zhang B, Jiang H, Wu J, et al. m6A demethylase FTO attenuates cardiac dysfunction by regulating glucose uptake and glycolysis in mice with pressure overload-induced heart failure. Signal Transduct Target Ther, 2021; 6(1): 377. doi: 10.1038/s41392-021-00699-w [91] Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-Dependent N(6)-Methyladenosine regulates cardiac function during remodeling and repair. Circulation, 2019; 139(4): 518-532. doi: 10.1161/CIRCULATIONAHA.118.033794 [92] Shen W, Li H, Su H, et al. FTO overexpression inhibits apoptosis of hypoxia/reoxygenation-treated myocardial cells by regulating m6A modification of Mhrt. Mol Cell Biochem, 2021; 476(5): 2171-2179. doi: 10.1007/s11010-021-04069-6 [93] Xu H, Wang Z, Chen M, et al. YTHDF2 alleviates cardiac hypertrophy via regulating Myh7 mRNA decoy. Cell Biosci, 2021; 11(1): 132. doi: 10.1186/s13578-021-00649-7 [94] Wang T, Tian J, Jin Y. VCAM1 expression in the myocardium is associated with the risk of heart failure and immune cell infiltration in myocardium. Sci Rep, 2021; 11(1): 19488. doi: 10.1038/s41598-021-98998-3 [95] NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1201 population-representative studies with 104 million participants. Lancet, 2021; 398(10304): 957-980. doi: 10.1016/S0140-6736(21)01330-1 [96] Wu Q, Yuan X, Han R, et al. Epitranscriptomic mechanisms of N6-methyladenosine methylation regulating mammalian hypertension development by determined spontaneously hypertensive rats pericytes. Epigenomics, 2019; 11(12): 1359-1370. doi: 10.2217/epi-2019-0148 [97] Zeng Y, Huang T, Zuo W, et al. Integrated analysis of m(6)A mRNA methylation in rats with monocrotaline-induced pulmonary arterial hypertension. Aging (Albany NY), 2021; 13(14): 18238-18256. [98] Hu L, Wang J, Huang H, et al. YTHDF1 regulates pulmonary hypertension through translational control of MAGED1. Am J Respir Crit Care Med, 2021; 203(9): 1158-1172. doi: 10.1164/rccm.202009-3419OC [99] Qin Y, Qiao Y, Li L, et al. The m(6)A methyltransferase METTL3 promotes hypoxic pulmonary arterial hypertension. Life Sci, 2021; 274: 119366. doi: 10.1016/j.lfs.2021.119366 [100] Zhou X L, Huang F J, Li Y, et al. SEDT2/METTL14-mediated m6A methylation awakening contributes to hypoxia-induced pulmonary arterial hypertension in mice. Aging (Albany NY), 2021; 13(5): 7538-7548. [101] Su H, Wang G, Wu L, et al. Transcriptome-wide map of m(6) A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics, 2020; 21(1): 39. doi: 10.1186/s12864-020-6462-y [102] Xu S, Xu X, Zhang Z, et al. The role of RNA m(6)A methylation in the regulation of postnatal hypoxia-induced pulmonary hypertension. Respir Res, 2021; 22(1): 121. doi: 10.1186/s12931-021-01728-6 [103] He Y, Xing J, Wang S, et al. Increased m6A methylation level is associated with the progression of human abdominal aortic aneurysm. Ann Transl Med, 2019; 7(24): 797. doi: 10.21037/atm.2019.12.65 [104] Li T, Wang T, Jing J, et al. Expression pattern and clinical value of key m6A RNA modification regulators in abdominal aortic aneurysm. J Inflamm Res, 2021; 14: 4245-4258. doi: 10.2147/JIR.S327152 [105] Zhong L, He X, Song H, et al. METTL3 induces AAA development and progression by modulating N6-methyladenosinedependent primary miR34a processing. Mol Ther Nucleic Acids, 2020; 21: 394-411. doi: 10.1016/j.omtn.2020.06.005 [106] Ma D, Liu X, Zhang J J, et al. Vascular smooth muscle FTO promotes aortic dissecting aneurysms via m6A modification of Klf5. Front Cardiovasc Med, 2020; 7: 592550. doi: 10.3389/fcvm.2020.592550 [107] Ju W, Liu K, Ouyang S, et al. Changes in N6-Methyladenosine modification modulate diabetic cardiomyopathy by reducing myocardial fibrosis and myocyte hypertrophy. Front Cell Dev Biol, 2021, 9: 702579. doi: 10.3389/fcell.2021.702579 [108] Yu Y, Pan Y, Fan Z, et al. LuHui derivative, a novel compound that inhibits the fat mass and obesity-associated (FTO), alleviates the inflammatory response and injury in hyperlipidemia-induced cardiomyopathy. Front Cell Dev Biol, 2021; 9: 731365. doi: 10.3389/fcell.2021.731365 [109] Dubey P K, Patil M, Singh S, et al. Increased m6A-RNA methylation and FTO suppression is associated with myocardial inflammation and dysfunction during endotoxemia in mice. Mol Cell Biochem, 2022; 477(1): 129-141. doi: 10.1007/s11010-021-04267-2 [110] Mo X B, Lei S F, Zhang Y H, et al. Detection of m(6)A-associated SNPs as potential functional variants for coronary artery disease. Epigenomics, 2018; 10(10): 1279-1287. doi: 10.2217/epi-2018-0007 [111] Mo X, Lei S, Zhang Y, et al. Genome-wide enrichment of m(6) A-associated single-nucleotide polymorphisms in the lipid loci. Pharmacogenomics J, 2019; 19(4): 347-357. doi: 10.1038/s41397-018-0055-z [112] Mo X B, Lei S F, Zhang Y H, et al. Examination of the associations between m(6)A-associated single-nucleotide polymorphisms and blood pressure. Hypertens Res, 2019; 42(10): 1582-1589. doi: 10.1038/s41440-019-0277-8 [113] Zhang H, Shi X, Huang T, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res, 2020; 48(11): 6251-6264. doi: 10.1093/nar/gkaa347 [114] Xiong X, Hou L, Park Y P, et al. Genetic drivers of m(6)A methylation in human brain, lung, heart and muscle. Nat Genet, 2021; 53(8): 1156-1165. doi: 10.1038/s41588-021-00890-3 [115] Zhou Y, Fang Y, Dai C, et al. PiRNA pathway in the cardiovascular system: a novel regulator of cardiac differentiation, repair and regeneration. J Mol Med (Berl), 2021; 99(12): 1681-1690. doi: 10.1007/s00109-021-02132-9 [116] Chen J, Liu Z, Ma L, et al. Targeting epigenetics and non-coding RNAs in myocardial infarction: from mechanisms to therapeutics. Front Genet, 2021; 12: 780649. doi: 10.3389/fgene.2021.780649 [117] Jakobi T, Siede D, Eschenbach J, et al. Deep characterization of circular RNAs from human cardiovascular cell models and cardiac tissue. Cells, 2020; 9(7): 1616. doi: 10.3390/cells9071616 [118] Li M, Parker B L, Pearson E, et al. Core functional nodes and sex-specific pathways in human ischaemic and dilated cardiomyopathy. Nat Commun, 2020; 11(1): 2843. doi: 10.1038/s41467-020-16584-z [119] Cui H, Chen Y, Li K, et al. Untargeted metabolomics identifies succinate as a biomarker and therapeutic target in aortic aneurysm and dissection. Eur Heart J, 2021; 42(42): 4373-4385. doi: 10.1093/eurheartj/ehab605 [120] Yan X, Jin J, Su X, et al. Intestinal flora modulates blood pressure by regulating the synthesis of intestinal-derived corticosterone in high salt-induced hypertension. Circ Res, 2020; 126(7): 839-853. doi: 10.1161/CIRCRESAHA.119.316394 [121] Barallobre-Barreiro J, Radovits T, Fava M, et al. Extracellular matrix in heart failure: role of ADAMTS5 in proteoglycan remodeling. Circulation, 2021; 144(25): 2021-2034. doi: 10.1161/CIRCULATIONAHA.121.055732 [122] Yokota T, McCourt J, Ma F, et al. Type V collagen in scar tissue regulates the size of scar after heart injury. Cell, 2020; 182(3): 545-562. doi: 10.1016/j.cell.2020.06.030 [123] Porritt R A, Zemmour D, Abe M, et al. NLRP3 inflammasome mediates immune-stromal interactions in vasculitis. Circ Res, 2021; 129(9): e183-e200. [124] Asp M, Giacomello S, Larsson L, et al. A spatiotemporal organwide gene expression and cell atlas of the developing human heart. Cell, 2019; 179(7): 1647-1660. doi: 10.1016/j.cell.2019.11.025 [125] Sikorski V, Karjalainen P, Blokhina D, et al. Epitranscriptomics of ischemic heart disease-the IHD-EPITRAN study design and objectives. Int J Mol Sci, 2021; 22(12): 6630. doi: 10.3390/ijms22126630 -


 投稿系统
投稿系统


 下载:
下载: