Altered expression profile of long non-coding RNAs during heart aging in mice
doi: 10.2478/fzm-2022-0015
-
Abstract:
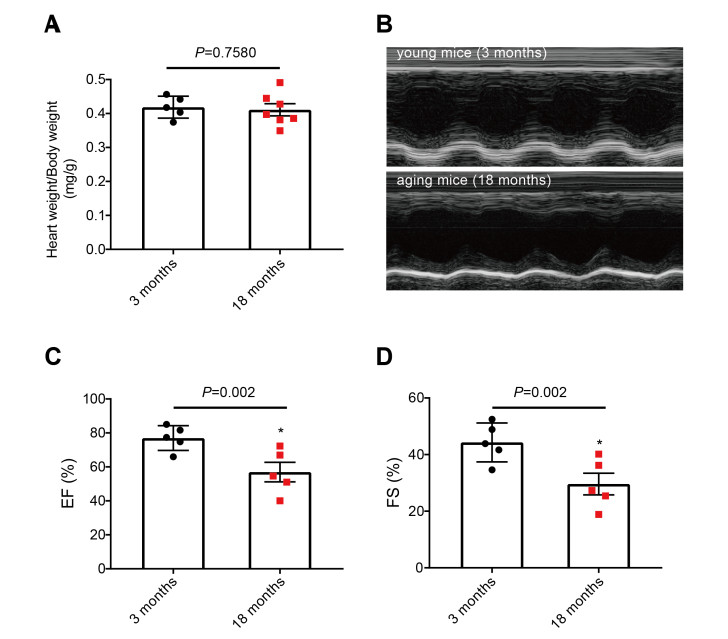
Objective Long noncoding RNAs (lncRNAs) play an important role in regulating the occurrence and development of cardiovascular diseases. However, the role of lncRNAs in heart aging remains poorly understood. The objective of this study was to identify differentially expressed lncRNAs in the heart of aging mice and elucidate the relevant regulatory pathways of cardiac aging. Materials and methods Echocardiography was used to detect the cardiac function of 18-months (aged) and 3-months (young) old C57BL/6 mice. Microarray analysis was performed to unravel the expression profiles of lncRNAs and mRNAs, and qRT-PCR to verify the highly dysregulated lncRNAs. Results Our results demonstrated that the heart function in aged mice was impaired relative to young ones. Microarray results showed that 155 lncRNAs were upregulated and 37 were downregulated, and 170 mRNAs were significantly upregulated and 44 were remarkably downregulated in aging hearts. Gene ontology analysis indicated that differentially expressed genes are mainly related to immune function, cell proliferation, copper ion response, and cellular cation homeostasis. KEGG pathway analysis showed that the differentially expressed mRNAs are related to cytokine-cytokine receptor interaction, inflammatory mediator regulation of TRP channels, and the NF-kappa B signaling pathway. Conclusion These results imply that the differentially expressed lncRNAs may regulate the development of heart aging. This study provides a new perspective on the potential effects and mechanisms of lncRNAs in heart aging. -
Key words:
- heart aging /
- long noncoding RNAs /
- gene microarray /
- expression profile /
- cold stress /
- cardiovascular diseases
-
Figure 2. Differential expression of lncRNAs and mRNAs in cardiac tissues from aged and young mice
Hierarchical clustering and volcano plots of data from microarray analysis showing the differences in lncRNA (A, C) and mRNA (B, D) expression profiles in the hearts of aged mice relative to young mice. qRT-PCR verification of the upregulated lncRNAs (E) and downregulated lncRNAs (F).
Figure 3. GO analysis of differentially expressed genes for their potential roles in regulating biological process (BP), cellular component (CC) and molecular function (MF)
The biological process of top ten upregulated (A) and downregulated (B) mRNAs predicted by Go analysis. The analysis of the cellular component for the upregulated (C) and downregulated (D) genes. (E and F) The predicted molecular function of upregulated and downregulated mRNAs, respectively.
Table 1. Ten most up- and downregulated lncRNA in the senile mice compared to the young mice tissues by volcano plot
Seqname Gene Symbol Fold Change P-Value False discovery rate Chromosome Relationship Regulation uc007pgi.1 Igh-A 16.4211129 0.0123091017135 0.704002480 chr12 intergenic up uc009cfw.1 M34473 11.9186002 0.0120988638993 0.703893166 chr6 intergenic up ENSMUST00000162347 Gm16028 11.4247018 0.0002546295660 0.324552338 chr1 natural antisense up ENSMUST00000143329 Pak7 11.2164783 0.0005184195769 0.352156281 chr2 exon sense-overlapping up uc007bte.1 BC038927 9.8736198 0.0008021783571 0.389972717 chr1 intergenic up uc029tuy.1 BC038927 9.0601077 0.0002724285562 0.324552338 chr1_GL456221_random exon sense-overlapping up uc029qqx.1 BC038927 8.9514910 0.0003940837912 0.335359940 chr1 intergenic up ENSMUST00000127039 Cpxm2 8.6573415 0.0316924115364 0.835551190 chr7 exon sense-overlapping up uc007bts.1 BC038927 8.5146551 0.0005341530809 0.352156281 chr1 intergenic up ENSMUST00000149852 Myh7 8.0777575 0.0031840612119 0.578467076 chr14 exon sense-overlapping up AK047207 AK047207 18.5809419 0.0541673031371 0.90042180 chr4_GL456350_random intergenic down TCONS_00008432 XLOC_006834 11.1899786 0.0096746130928 0.673876406 chr13 intergenic down NR_045178 1700042O10Rik 10.1800255 0.0002896109249 0.324552338 chr11 intronic antisense down ENSMUST00000146359 Ddc 9.2777667 0.0055688768544 0.620442862 chr11 exon sense-overlapping down ENSMUST00000180853 B430219N15Rik 5.1778340 0.0038964965731 0.612011531 chr10 natural antisense down uc029usn.1 Gm5859 4.7961460 0.0427001224829 0.890663465 chr4 intergenic down ENSMUST00000147230 B430219N15Rik 4.7234781 0.0019670790981 0.521930155 chr10 natural antisense down ENSMUST00000149439 Wdr72 4.6269702 0.0002775462694 0.324552338 chr9 exon sense-overlapping down ENSMUST00000168048 Gm17171 3.5947210 0.0127512174729 0.713277281 chr2 intronic antisense down ENSMUST00000095448 E230001N04Rik 3.5589724 0.0179762591263 0.759308574 chr17 intergenic down Table 2. Ten most up- and downregulated mRNA in the senile mice compared to the young mice tissues by volcano plot
Gene Symbol Fold Change Regulation P-Value FDR Chromosome Cyp2b10 19.4177580 up 0.0012091 0.368406446 cytochrome P450, family 2, subfamily b, polypeptide 10 Klk1b21 18.1814910 up 0.0023379 0.420447958 kallikrein 1-related peptidase b21 Klk1b16 16.9975930 up 0.0005335 0.306936746 kallikrein 1-related peptidase b16 Klk1b27 16.867272 up 0.0030525 0.435434485 kallikrein 1-related peptidase b27 Ccl8 16.647476 up 0.0030389 0.435434485 chemokine (C-C motif) ligand 8 Klk1b26 16.595657 up 0.0004407 0.306936746 kallikrein 1-related petidase b26 Klk1b8 14.5656130 up 0.0002850 0.306936746 kallikrein 1-related peptidase b8 Cyp2b9 13.870040 up 0.0064780 0.505295448 cytochrome P450, family 2, subfamily b, polypeptide 9 Klk1b22 13.5142910 up 0.0009369 0.339547062 kallikrein 1-related peptidase b22 Egfbp2 12.8234330 up 0.0003858 0.306936746 epidermal growth factor binding protein type B Pkp1 8.5662940 down 0.0363041 0.727009964 plakophilin 1 2210407C18Rik 4.6002815 down 0.0284802 0.704663955 RIKEN cDNA 2210407C18 gene Lsp1 4.4957683 down 0.0001650 0.231154293 lymphocyte specific 1 Kcne1 3.8354920 down 0.0019954 0.398112975 potassium voltage-gated channel, Isk-related subfamily, member 1 Bcat1 3.7411754 down 0.0393461 0.727009964 branched chain aminotransferase 1, cytosolic Wdr72 3.5439451 down 0.0005649 0.306936746 WD repeat domain 72 Per2 3.4188627 down 0.0241152 0.684321395 period circadian clock 2 Lrat 3.3565073 down 0.0347539 0.725192325 lecithin-retinol acyltransferase(phosphatidylcholine-retinol-O-acyltransferase) Med13l 3.1438580 down 0.0108414 0.571936897 mediator complex subunit 13-like Tmem100 2.9812499 down 0.0162492 0.625508937 transmembrane protein 100 Table 3. Five up-regulated and four down-regulated lncRNA in the aging mice heart compared to the young mice tissues by qRT-PCR
Sample GAPDH (Ct) uc007pgi.1 (Ct) uc009cfw.1 (Ct) ENSMUST00000 143329(Ct) UC007bte.1 (Ct) ENSMUST00000 162347(Ct) ENSMUST00000 146359(Ct) NR-045178 (Ct) TCONS-00008432 (Ct) ENSMUST00000 180853(Ct) Young 17.6018 30.5075 31.4878 32.7336 28.7084 30.8973 32.8515 36.9808 35.9523 Undetermined Young 24.9457 29.6875 33.6435 Undetermined 29.4312 28.5655 31.4305 30.6550 31.8039 34.1035 Young 18.6359 28.4475 29.0027 31.6887 28.7353 30.6615 30.5467 28.8232 31.9750 32.0288 Young 21.7831 27.7026 Undetermined 27.1878 27.2907 29.4938 30.2939 27.3035 31.2758 35.9406 Young 18.1955 27.9258 28.9289 27.3102 26.4704 30.8142 27.2946 33.1586 Old 16.9513 28.9224 30.3551 29.5674 29.8970 31.4611 37.1237 33.0738 Undetermined 30.8844 Old 21.2262 27.4433 35.9324 28.7717 26.1900 28.4325 Undetermined Undetermined Undetermined 35.0848 Old 16.6436 28.2656 29.3889 35.8320 28.1946 30.7540 36.5523 31.6621 34.8063 30.3491 Old 16.8656 30.4991 Undetermined 37.0713 25.5742 30.0247 Undetermined Undetermined Undetermined 32.0592 Old 17.4761 27.3305 34.9349 30.2778 25.4458 Undetermined 35.6645 36.9855 -
[1] Khaltourina D, Matveyev Y, Alekseev A, et al. Aging fits the disease criteria of the international classification of diseases. Mech Ageing Dev, 2020; 189(366): 111230. [2] Childs B G, Baker D J, Wijshake T, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science, 2016; 354(6311): 472-477. doi: 10.1126/science.aaf6659 [3] Lakatta E G. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part Ⅲ: cellular and molecular clues to heart and arterial aging. Circulation, 2003; 107(3): 490-497. doi: 10.1161/01.CIR.0000048894.99865.02 [4] Lakatta E G, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part Ⅱ: the aging heart in health: links to heart disease. Circulation, 2003; 107(2): 346-354. doi: 10.1161/01.CIR.0000048893.62841.F7 [5] Shih H, Lee B, Lee R J, et al. The aging heart and post-infarction left ventricular remodeling. J Am Coll Cardiol, 2011; 57(1): 9-17. doi: 10.1016/j.jacc.2010.08.623 [6] Torella D, Rota M, Nurzynska D, et al. Cardiac stem cell and myocyte aging, heart failure, and insulin-like growth factor-1 overexpression. Circ Res, 2004; 94(4): 514-524. doi: 10.1161/01.RES.0000117306.10142.50 [7] Zhao C, Li G, Li J. Non-coding RNAs and cardiac aging. Adv Exp Med Biol, 2020; 1229: 247-258. [8] Wang X, Jiang Y, Bai Y, et al. Association between air temperature and the incidence of acute coronary heart disease in northeast China. Clin Interv Aging, 2020; 15: 47-52. doi: 10.2147/CIA.S235941 [9] Liang J, Yin K, Cao X, et al. Attenuation of low ambient temperatureinduced myocardial hypertrophy by atorvastatin via promoting Bcl-2 expression. Cell Physiol Biochem, 2017; 41(1): 286-295. doi: 10.1159/000456111 [10] Li H M, Liu X, Meng Z Y, et al. Kanglexin delays heart aging by promoting mitophagy. Acta Pharmacol Sin, 2021, Online ahead of print. [11] Li J, Li S H, Dong J, et al. Long-term repopulation of aged bone marrow stem cells using young Sca-1 cells promotes aged heart rejuvenation. Aging Cell, 2019; 18: e13026. [12] Li Q, Liu X, Wei J. Ageing related periostin expression increase from cardiac fibroblasts promotes cardiomyocytes senescent. Biochem Biophys Res Commun, 2014; 452(3): 497-502. doi: 10.1016/j.bbrc.2014.08.109 [13] Moslehi J, DePinho R A, Sahin E. Telomeres and mitochondria in the aging heart. Circ Res, 2012; 110(9): 1226-1237. doi: 10.1161/CIRCRESAHA.111.246868 [14] Petrosillo G, Matera M, Moro N, et al. Mitochondrial complex I dysfunction in rat heart with aging: critical role of reactive oxygen species and cardiolipin. Free Radic Biol Med, 2009; 46(1): 88-94. doi: 10.1016/j.freeradbiomed.2008.09.031 [15] Martin-Fernandez B, Gredilla R. Mitochondria and oxidative stress in heart aging. Age (Dordr), 2016; 38(4): 225-238. doi: 10.1007/s11357-016-9933-y [16] Ramos-Marques E, Garcia-Mendivil L, Perez-Zabalza M, et al. Chronological and biological aging of the human left ventricular myocardium: analysis of microRNAs contribution. Aging Cell, 2021; 20(7): e13383. [17] Chen K, Wang S, Sun Q W, et al. Klotho deficiency causes heart aging via impairing the Nrf2-GR pathway. Circ Res, 2021; 128(4): 492-507. doi: 10.1161/CIRCRESAHA.120.317348 [18] Wen D T, Zheng L, Li J X, et al. The activation of cardiac dSir2- related pathways mediates physical exercise resistance to heart aging in old Drosophila. Aging (Albany NY), 2019; 11(17): 7274-7293. [19] Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 2009; 458(7235): 223-227. doi: 10.1038/nature07672 [20] Guttman M, Russell P, Ingolia NT, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell, 2013; 154(1): 240-251. doi: 10.1016/j.cell.2013.06.009 [21] Chen Y, Zhou X, Huang C, et al. LncRNA PART1 promotes cell proliferation and progression in non-small-cell lung cancer cells via sponging miR-17-5p. J Cell Biochem, 2020; 122(3-4): 315-325. [22] Chen H, Chen J. LncRNA SOX21-AS1 promotes the growth and invasiveness of osteosarcoma cells through miR-7-5p/IRS2 regulatory network. Arch Med Res, 2020; 52(3): 294-303. [23] Chen X J, An N. Long noncoding RNA ATB promotes ovarian cancer tumorigenesis by mediating histone H3 lysine 27 trimethylation through binding to EZH2. J Cell Mol Med, 2021; 25(1): 37-46. doi: 10.1111/jcmm.15329 [24] Jae N, Heumuller A W, Fouani Y, et al. Long non-coding RNAs in vascular biology and disease. Vascul Pharmacol, 2019; 114(3): 13-22. [25] Xie Z, Xia W, Hou M. Long intergenic noncoding RNAp21 mediates cardiac senescence via the Wnt/betacatenin signaling pathway in doxorubicin-induced cardiotoxicity. Mol Med Rep, 2018; 17(2): 2695-2704. [26] Cabiati M, Sapio A, Salvadori C, et al. Evaluation of transcriptional levels of the natriuretic peptides, endothelin-1, adrenomedullin, their receptors and long non-coding RNAs in rat cardiac tissue as cardiovascular biomarkers of aging. Peptides, 2020; 123: 170173. doi: 10.1016/j.peptides.2019.170173 [27] Abdelmohsen K, Panda A, Kang M J, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell, 2013; 12(5): 890-900. doi: 10.1111/acel.12115 [28] Lai C H, Chen A T, Burns A B, et al. RAMP2-AS1 regulates endothelial homeostasis and aging. Front Cell Dev Biol, 2021; 9: 635307. doi: 10.3389/fcell.2021.635307 [29] Zheng Y, Liu T, Li Q, et al. Integrated analysis of long non-coding RNAs (lncRNAs) and mRNA expression profiles identifies lncRNA PRKG1-AS1 playing important roles in skeletal muscle aging. Aging (Albany NY), 2021; 13: 15044-15060. [30] Hao K, Lei W, Wu H, et al. LncRNA-Safe contributes to cardiac fibrosis through Safe-Sfrp2-HuR complex in mouse myocardial infarction. Theranostics, 2019; 9(24): 7282-7297. doi: 10.7150/thno.33920 [31] Cai B, Ma W, Wang X, et al. Targeting LncDACH1 promotes cardiac repair and regeneration after myocardium infarction. Cell Death Differ, 2020; 27(7): 2158-2175. doi: 10.1038/s41418-020-0492-5 [32] Wang J, Zhang S, Li X, et al. LncRNA SNHG7 promotes cardiac remodeling by upregulating ROCK1 via sponging miR-34-5p. Aging (Albany NY), 2020; 12(11): 10441-10456. [33] Chen J, Zou Q, Lv D, et al. Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget, 2018; 9(2): 1524-1541. doi: 10.18632/oncotarget.23290 [34] Greenig M, Melville A, Huntley D, et al. Cross-sectional transcriptional analysis of the aging murine heart. Front Mol Biosci, 2020; 7: 565530. doi: 10.3389/fmolb.2020.565530 [35] Yu W, Mengersen K, Wang X, et al. Daily average temperature and mortality among the elderly: a meta-analysis and systematic review of epidemiological evidence. Int J Biometeorol, 2012; 56(4): 569-581. doi: 10.1007/s00484-011-0497-3 [36] von Klot S, Zanobetti A, Schwartz J. Influenza epidemics, seasonality, and the effects of cold weather on cardiac mortality. Environ Health, 2012; 11(1): 74. doi: 10.1186/1476-069X-11-74 [37] Cong P, Liu Y, Liu N, et al. Cold exposure induced oxidative stress and apoptosis in the myocardium by inhibiting the Nrf2-Keap1 signaling pathway. BMC Cardiovasc Disord, 2018; 18(1): 36. doi: 10.1186/s12872-018-0748-x [38] Dowery R, Benhamou D, Benchetrit E, et al. Peripheral B-cells repress B-cell regeneration in aging through a TNFalpha/IGFBP-1/IGF1 immune-endocrine axis. Blood, 2021, 138(19): 1817-1829. doi: 10.1182/blood.2021012428 -


 投稿系统
投稿系统


 下载:
下载: