Circulating CCRR serves as potential novel biomarker for predicting acute myocardial infarction
doi: 10.1515/fzm-2024-0015
-
Abstract:
Objective Cold regions exhibit a high prevalence of cardiovascular disease, particularly acute myocardial infarction (AMI), which is one of the leading causes of death associated with cardiovascular conditions. Cardiovascular disease is closely linked to the abnormal expression of long non-coding RNA (lncRNA). This study investigates whether circulating levels of lncRNA cardiac conduction regulatory RNA (CCRR) could serve as a biomarker for AMI. Materials and methods We measured circulating CCRR from whole blood samples collected from 68 AMI patients and 69 non-AMI subjects. An AMI model was established using C57BL/6 mice. Quantitative reverse transcription PCR (qRT-PCR) was used to assess CCRR expression. Exosomes were isolated from cardiomyocytes, and their characteristics were evaluated using electron microscope and nanoparticle tracking analysis. The exosome inhibitor GW4869 was employed to examine the effect of exosomal CCRR on cardiac function using echocardiography. Protein expression was detected using Western blot and immunofluorescence staining. Results The circulating level of CCRR was significantly higher in AMI patients (1.93 ± 0.13) than in nonAMI subjects (1.00 ± 0.05, P < 0.001). The area under the ROC curve (AUC) of circulating CCRR was 0.821. Similar changes in circulating CCRR levels were consistently observed in an AMI mouse model. Exosomal CCRR derived from hypoxia-induced cardiomyocytes and cardiac tissue after AMI were increased, a change that was reversed by GW4869. Additionally, CCRR-overexpressing exosomes improved cardiac function in AMI. Conclusion Circulating lncRNA CCRR is a potential predictor of AMI. Exosomal CCRR plays a role in the communication between the heart and other organs through circulation. -
Key words:
- acute myocardial infarction /
- lncRNA /
- cardiac conduction regulatory RNA /
- exosome
-
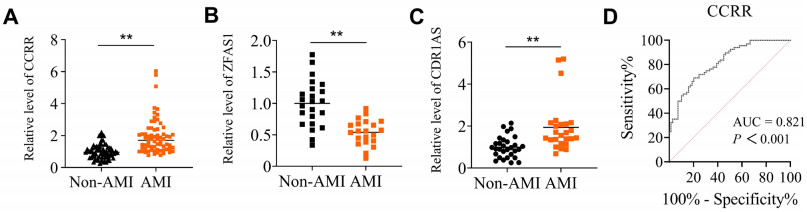
Figure 1. Analyses of CCRR levels in whole blood of acute myocardial infarction (AMI) patients
(A) The expression levels of lncRNA CCRR in whole blood samples from AMI patients and non-AMI subjects were comparatively analyzed using qRT-PCR. N = 68 for AMI and N = 69 for non-AMI subjects. (B and C) Circulating levels of lncRNA ZFAS1 (B) and CDR1AS (C). The expression levels of lncRNA ZFAS1 in whole blood samples; N = 21 for AMI and N = 23 for non-AMI subjects. The expression levels of lncRNA CDR1AS in whole blood samples; N = 25 for AMI and N = 30 for non-AMI subjects. Data are represented as means ± SEM; **P < 0.01 vs. Non-AMI. (D) The area under the ROC curve (AUC) was analyzed to determine the predictive power of circulating lncRNA levels for AMI using non-AMI subjects as control.
Figure 2. The expression of lncRNA CCRR in ischemic border zone of the heart and whole blood after acute myocardial infarction (AMI) surgery at different time points
(A, B) Changes of CCRR in cardiac tissues (A) and blood (B) CCRR levels in a mouse model of AMI at various time points (2 h, 6 h, 12 h and 24 h). The number of cardiac tissue sample was: N = 20 for sham, N = 12 for AMI 2 h, N = 14 for AMI 6 h, N = 14 for AMI 12 h, N = 8 for AMI 24 h. The number of blood sample was: N = 27 for sham, N = 11 for AMI 2h, N = 11 for AMI 6 h, N = 15 for AMI 12 h, N = 15 for AMI 24 h. AMI represent acute myocardial infarction with LAD. Data are represented as means ± SEM; *P < 0.05 vs. Sham, **P < 0.01 vs. Sham.
Figure 3. Exosomes as new vesicular lipid transporters involved in changes of CCRR in whole blood after acute myocardial infarction (AMI)
(A) GW4869 inhibited CD63 expression in the heart of AMI mice. N = 6 per group. (B) GW4869 inhibits CD63 expression in the cytoplasm of cardiomyocytes in GWAMI mice. CD63 was marked by red colour, α-actinin by green colour and DAPI by blue colour. Scale bar: 20 μm. N = 3 per group. (C) Confocal colocalization analysis of CCRR (red) and CD63 (green) in primary cultured neonatal mouse cardiomyocytes. CCRR was marked by red colour, CD63 by green colour and DAPI by blue colour. Scale bar: 5 μm. (D) Confocal co-localization analysis of CCRR and CD63 in mouse myocardium. CCRR was marked by red colour, CD63 by green colour and DAPI by blue colour. Scale bar: 20 μm. (E, F) CCRR levels in myocardial tissue (E) of mice and whole blood (F) after AMI mice were treated by GW4869. N = 16 per group. Data are represented as means ± SEM; *P < 0.05 vs. Sham, **P < 0.01 vs. Sham, #P < 0.05 vs. AMI, ##P < 0.01 vs. AMI.
Figure 4. Characterization of cardiomyocytes-derived exosomes
(A) Exosomes were extracted from cardiomyocyte supernatants under normoxic or hypoxic conditions. (B) Electron micrograph-analyzed cardiomyocyte exosomes. Scale bar: 200 nmol/L. (C) The size distribution of nor-Exo and hypo-Exo determined by nanoparticle tracking analysis (NTA). (D) Western blot of exosomal marker expression in cardiomyocyte exosomes. (E) Primary neonatal mouse cardiomyocytes were cultured with PKH26-labeled exosomes or without PKH26-labeled exosomes at 37℃, 5% CO2 for 24 h. PKH26 was marked by red colour, α-actinin by green colour and DAPI by blue colour. Scale bar: 10 μm.
Figure 5. GW4869 treatment mitigates myocardial infarction-induced cardiac dysfunction
(A) Flowchart of in vivo experimental design. (B) Western blot showing the protein level of CD63 and CD81 in Sham control mice cardiac exosomes (ShamExo), AMI mice cardiac exosomes (AMI-Exo) and GW4869-AMI mice cardiac exosomes N = 3. (C) lncRNA CCRR levels in the cardiac exosomes. N = 7 per group. (D) Representative echocardiographic images showing heart function among the different groups in the 12 h following AMI. (E-H) Quantitative analysis of left ventricular ejection fraction (E) left ventricular fraction shortening (F) left ventricular end-diastolic diameter (G) and left ventricular systolic diameter (H) among the different groups. N = 4 mice per group. Data are represented as means ± SEM; *P < 0.05 vs. Sham, **P < 0.01 vs. Sham, ***P < 0.001 vs. Sham, #P < 0.05 vs. AMI, ##P < 0.01 vs. AMI, ###P < 0.001 vs. AMI.
Figure 6. Administration of CCRR overexpressed exosomes improves cardiac 3 function in acute myocardial infarction mice
(A) Flowchart of in vivo experimental design. (B) Immunofluorescence images of cardiac tissue after intravenous injection of PKH26-labeled exosomes or PBS in mice. Scale bar: 20 μm. (C) Expression level of lncRNA CCRR in exosomes. N = 6 per group. (D) lncRNA CCRR levels in myocardial tissue. N = 7-8 per group. (E) Representative echocardiographic images showing cardiac function among the different groups on the 12 h following AMI. (F-I) Quantitative analysis of left ventricular ejection fraction (F) left ventricular fraction shortening (G) left ventricular end-diastolic diameter (H) and left ventricular systolic diameter (I) among the different groups. N = 6-8 mice per group. Data are represented as means ± SEM; *P < 0.05 vs. Sham, **P < 0.01 vs. Sham, #P < 0.05 vs. NC-Exos-AMI; ##P < 0.01 vs. NC-Exos-AMI.
Figure 7. Schematic illustration of delivery of the exosomal lncRNA CCRR into circulation in acute myocardial infarction injury. Exosomal lncRNA CCRR derived from hypoxic cardiomyocytes is released into circulation. Circulating exosomal lncRNA CCRR may serves as a promising novel biomarker for AMI risk prediction.
Table 1. Basic characteristic indicators and diagnostic indicators of non-AMI control subjects and myocardial infarction patients
Characteristics Statistical items Non-AMI control AMI P value Age N (Missing) 68 (1) 68 0.5671 Mean (SEM) 61.57 (1.08) 60.60 (1.30) Min, Max 40.00, 80.00 42.00, 82.00 Median 61.50 60.50 Range 56.00-68.00 52.00-69.00 Gender Male 33 (47.83%) 43 (63.24%) 0.0705 Female 36 (52.17%) 25 (36.76%) Total (Missing) 69 68 Hypertension Yes 35 (55.56%) 30 (44.12%) 0.1936 No 28 (44.44%) 38 (55.88%) Total (Missing) 63 (6) 68 Smoking Yes 20 (31.75%) 30 (44.12%) 0.1475 No 43 (68.25%) 38 (55.88%) Total (Missing) 63 (6) 68 Diabetes Yes 17 (26.98%) 19 (27.94%) 0.9034 No 46 (73.02%) 49 (72.06%) Total (Missing) 63 (6) 68 HDL N (Missing) 62 (7) 67 (1) 0.3831 Mean (SEM) 1.1050 (0.0336) 1.7870 (0.7482) Min, Max 0.530, 1.820 0.540, 51.120 Median 1.095 1.040 Range 0.9075-1.2550 0.860-1.230 LDL N (Missing) 62 (7) 67 (1) 0.0252 Mean (SEM) 2.700 (0.1125) 3.059 (0.1115) Min, Max 1.160, 4.870 1.520, 6.420 Median 2.580 2.860 Range 2.155-3.225 2.390-3.570 CHOL N (Missing) 62 (7) 67 (1) 0.0158 Mean (SEM) 4.368 (0.1483) 4.900 (0.1577) Min, Max 2.340, 8.720 2.760, 10.310 Median 4.245 4.740 Range 3.540-5.135 4.030-5.620 TG N (Missing) 62 (7) 67 (1) 0.4304 Mean (SEM) 1.932 (0.1736) 2.123 (0.1681) Min, Max 0.490, 7.560 0.540, 6.090 Median 1.620 1.670 Range 1.058-2.205 1.030-3.000 AST N (Missing) 15 (54) 49 (19) 0.0011 Mean (SEM) 26.43 (2.668) 181.7 (24.90) Min, Max 17.00, 53.00 20.00, 769.00 Median 23.50 85.00 Range 19.00-28.00 41.00-287.30 LDH N (Missing) 6 (63) 59 (9) 0.0514 Mean (SEM) 226.9 (52.04) 918.5 (110.1) Min, Max 136.50, 484.00 148.00, 5437.0 Median 184.70 674.00 Range 170.60-263.20 391.00-1094.0 CK-MB N (Missing) 30 (39) 39 (29) 0.0011 Mean (SEM) 1.042 (0.1597) 71.63 (18.15) Min, Max 0.200, 4.500 0.300, 576.20 Median 0.720 27.60 Range 0.630-1.300 1.900-106.30 CK N (Missing) 6 (63) 59 (9) 0.0436 Mean (SEM) 86.30 (13.90) 1522 (220.7) Min, Max 45.10, 136.90 33.00, 6779 Median 77.05 844.0 Range 60.18-121.6 178.0-2415 HBDH N (Missing) 5 (64) 35 (33) 0.0425 Mean (SEM) 118.7 (7.825) 638.5 (92.56) Min, Max 101.0, 139.5 134.8, 2367 Median 111.8 438.6 Range 103.6-137.4 265.4-724.4 cTn N (Missing) 31 (38) 48 (20) 0.0218 Mean (SEM) 6.682 (2.566) 21341 (7306) Min, Max 0.000, 70.70 2.900, 289880 Median 2.210 3727 Range 1.400, 5.100 178.2, 14289 AST: aspartate transaminase; CHOL: total cholesterol; CK: creatine kinase; CK-MB: creatine kinase MB; HBDH: hydroxybutyrate dehydrogenase; HDL: high density cholesterol; LDH: lactic dehydrogenase; LDL: low density cholesterol; TG: triglyceride. Table 2. The Statistical Analysis of Circulating lncRNA CCRR
lncRNA Statistical items Non-AMI control AMI P value CCRR N (Missing) 69 68 < 0.001 Mean (SEM) 1.0000 (0.0544) 1.9290 (0.1323) Min, Max 0.257, 2.214 0.768, 6.033 Range 0.649, 1.247 1.146, 2.292 Table 3. Univariate regression analysis for the association of CCRR with basic characteristic indicators between AMI patients and non-AMI control subject
Parameter Estimate SE Wald P value OR 95%CI lower upper CCRR 2.333 0.4527 26.543 < 0.0001 10.31 4.573 27.23 Age -0.0101 0.0176 0.3327 0.5635 0.9899 0.9561 1.025 Gender 0.6293 0.3483 3.265 0.0705 1.876 0.9524 3.744 Diabetes 0.0481 0.3919 0.0150 0.9024 1.049 0.4861 2.277 HDL 0.0599 0.0957 0.3916 0.3258 1.062 0.9581 2.055 LDL 0.4579 0.2093 4.787 0.0232 1.581 1.063 2.428 TG 0.1041 0.1316 0.6266 0.4247 1.110 0.8603 1.451 CHOL 0.3719 0.1594 5.448 0.0136 1.450 1.077 2.016 HDL: high density cholesterol; LDL: low density cholesterol; TG: triglyceride; CHOL: total cholesterol. Table 4. Multivariate regression analysis for the association of CCRR with basic characteristic indicators between AMI patients and non-AMI control subjects
Parameter Estimate SE Wald P value OR 95%CI lower upper CCRR 2.516 0.5112 24.22 < 0.0001 12.38 4.982 37.41 Age -0.0099 0.0244 0.1640 0.6855 0.9902 0.9433 1.039 Gender 0.7746 0.5167 2.247 0.1338 2.170 0.8018 6.183 Diabetes 0.4361 0.5437 0.6434 0.4225 1.547 0.5370 4.609 HDL 0.01978 0.0911 0.0471 0.8281 1.020 0.9174 2.026 LDL 0.2817 0.5466 0.2656 0.6063 1.325 0.3725 3.538 TG -0.0263 0.1997 0.0174 0.8951 0.974 0.6485 1.438 CHOL 0.4090 0.4215 0.9413 0.3319 1.505 0.7449 4.332 HDL: high density cholesterol; LDL: low density cholesterol; TG: triglyceride; CHOL: total cholesterol. -
[1] Roth G A, Mensah G A, Johnson C O, et al. Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J. Am. Coll. Cardiol, 2020; 76: 2982-3021. [2] Liu S, Li Y, Zeng X, et al. Burden of cardiovascular diseases in China, 1990-2016: findings from the 2016 global burden of disease study. JAMA Cardiology, 2019; 4 (4): 342-352. doi: 10.1001/jamacardio.2019.0295 [3] Zhao D. Epidemiological features of cardiovascular disease in Asia. JACC: Asia, 2021; 1 (1): 1-13. doi: 10.1016/j.jacasi.2021.04.007 [4] Li X, Wu C, Lu J, et al. Cardiovascular risk factors in China: a nationwide population-based cohort study. The Lancet Public Health, 2020; 5 (12): e672-e681. doi: 10.1016/S2468-2667(20)30191-2 [5] Lei J, Chen R, Yin P, et al. Association between cold spells and mortality risk and burden: a nationwide study in China. Environmental Health Perspectives, 2022; 130 (2): 027006. doi: 10.1289/EHP9284 [6] Schulte C, Barwari T, Joshi A, et al. Noncoding RNAs versus protein biomarkers in cardiovascular disease. Trends Mol Med, 2020; 26: 583-596. doi: 10.1016/j.molmed.2020.02.001 [7] Emdin M, Aimo A, Vergaro G, et al. sST2 predicts outcome in chronic heart failure beyond NT-proBNP and high-sensitivity troponin T. J Am Coll Cardiol, 2018; 72: 2309-2320. doi: 10.1016/j.jacc.2018.08.2165 [8] Wang Y, Sun X. The functions of LncRNA in the heart. Diabetes Res Clin Pract, 2020; 168: 108249. doi: 10.1016/j.diabres.2020.108249 [9] Mozaffarian D, Benjamin E J, Go A S, et al. Heart disease and stroke statistics-2015 update: a report from the American Heart Association. Circulation, 2015; 131: e29-322. doi: 10.1161/circ.131.suppl_2.o29 [10] Kopp F, Mendell J T. Functional classification and experimental dissection of long noncoding RNAs. Cell, 2018; 172: 393-407. doi: 10.1016/j.cell.2018.01.011 [11] Chang G, Zhang W, Zhang M, et al. Clinical value of circulating ZFAS1 and miR-590-3p in the diagnosis and prognosis of chronic heart failure. Cardiovasc. Toxicol, 2021; 21: 880-888. doi: 10.1007/s12012-021-09678-7 [12] Sharma S, Findlay G M, Bandukwala H S, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA protein scaffold complex. Proc Natl Acad Sc, 2011; 108: 11381-11386. doi: 10.1073/pnas.1019711108 [13] Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res, 2017; 120: 381-399. doi: 10.1161/CIRCRESAHA.116.308434 [14] Cao M, Luo H, Li D, et al. Research advances on circulating long noncoding RNAs as biomarkers of cardiovascular diseases. Int J Cardiol, 2022; 353: 109-117. doi: 10.1016/j.ijcard.2022.01.070 [15] Zhang Y, Sun L, Xuan L, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep, 2016; 6: 2384. [16] Wang X, Wang L, Ma Z, et al. Early expressed circulating long noncoding RNA CHAST is associated with cardiac contractile function in patients with acute myocardial infarction. Int J Cardiol, 2020; 302: 15-20. doi: 10.1016/j.ijcard.2019.12.058 [17] Tang Y, Zheng J, Sun Y, et al. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J, 2009; 50: 377-387. doi: 10.1536/ihj.50.377 [18] Gao L, Liu Y, Guo S, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell. Physiol. Biochem, 2017; 44: 1497-1508. doi: 10.1159/000485588 [19] Azat M, Huojiahemaiti X, Gao R, et al. Long noncoding RNA MIAT: a potential role in the diagnosis and mediation of acute myocardial infarction. Mol Med Rep, 2019; 20: 5216-5222. [20] Wang L, Wang L, Wang Q. Constitutive activation of the NEAT1/miR-22-3p/Ltb4r1 signaling pathway in mice with myocardial injury following acute myocardial infarction. Aging (Albany NY), 2021; 13: 15307-15319. [21] Li M, Wang Y F, Yang X C, et al. Circulating long noncoding RNA LIPCAR acts as a novel biomarker in patients with ST-segment elevation myocardial infarction. Med Sci Monit, 2018; 24: 5064-5070. doi: 10.12659/MSM.909348 [22] Yan L, Zhang Y, Zhang W, et al. lncRNA-NRF is a potential biomarker of heart failure after acute myocardial infarction. J Cardiovasc Transl Res, 2020; 13: 1008-1015. doi: 10.1007/s12265-020-10029-0 [23] Zhang Y, Sun L, Xuan L, et al. Long non-coding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nat Commun, 2018; 9: 4176. doi: 10.1038/s41467-018-06637-9 [24] Khodayi M, Khalaj-Kondori M, Hoseinpour F M A, et al. Plasma lncRNA profiling identified BC200 and NEAT1 lncRNAs as potential blood-based biomarkers for late-onset Alzheimer's disease. EXCLI J, 2022; 21: 772-785. [25] Isaac R, Reis F C G, Ying W, et al. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab, 2021; 33: 1744-1762. doi: 10.1016/j.cmet.2021.08.006 [26] Zheng M L, Liu X Y, Han R J, et al. Circulating exosomal long noncoding RNAs in patients with acute myocardial infarction. J Cell Mol Med, 2020; 24: 9388-9396. doi: 10.1111/jcmm.15589 [27] Catalano M, O'Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles, 2020; 9, 1703244. doi: 10.1080/20013078.2019.1703244 [28] Crea F. How epidemiology can improve the understanding of cardiovascular disease: from mechanisms to treatment. Eur Heart J, 2021; 42: 4503-4507. doi: 10.1093/eurheartj/ehab797 [29] Lerman B, Lerman L O. "Nothing burns like the cold": Cardiovascular disease in frigid zones. Frigid Zone Medicine, 2022; 2 (3): 129-131. doi: 10.2478/fzm-2022-0017 [30] Jin H. Increased risk of cardiovascular disease in cold temperatures. Frigid Zone Medicine, 2022; 2 (3): 138-139. doi: 10.2478/fzm-2022-0020 [31] Oitabén A, Fonseca P, Villanueva M J, et al. Emerging blood-based biomarkers for predicting immunotherapy response in NSCLC. Cancers (Basel), 2022; 14 (11), 2626. doi: 10.3390/cancers14112626 [32] Chaitman B R, Cyr D D, Alexander K P, et al. Cardiovascular and renal implications of myocardial infarction in the ISCHEMIA-CKD trial. Circ Cardiovasc Interv, 2022; 15: e012103. [33] Wang F, Li X, Xie X, et al. UCA1, a non-protein-coding RNA upregulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett, 2008; 582: 1919-1927. doi: 10.1016/j.febslet.2008.05.012 [34] Yan Y, Zhang B, Liu N, et al. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int, 2016; 2016: 8079372. [35] Zheng P F, Chen L Z, Liu P, et al. A novel lncRNA-miRNA-mRNA triple network identifies lncRNA XIST as a biomarker for acute myocardial infarction. Aging (Albany NY), 2022; 14: 4085-4106. [36] Wang Y, Zhao R, Liu W, et al. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longev, 2019; 2019: 7954657. [37] Gupta S, Knowlton A A. HSP60 trafficking in adult cardiac myocytes: role of the exosomal pathway. Am J Physiol Heart Circ Physiol, 2007; 292: H3052-3056. [38] King H W, Michael M Z, Gleadle J M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer, 2021; 12: 421. [39] Vicencio J M, Yellon D M, Sivaraman V, et al. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J Am Coll Cardiol, 2015; 65 (15): 1525-1536. [40] Tomasoni S, Longaretti L, Rota C, et al. Transfer of growth factor receptor mRNA via exosomes unravels the regenerative effect of mesenchymal stem cells. Stem Cells Dev, 2013; 22 (5): 772-778. [41] Jiao L, Li M, Shao Y, et al. lncRNA-ZFAS1 induces mitochondriamediated apoptosis by causing cytosolic Ca2+ overload in myocardial infarction mice model. Cell Death Dis, 2019; 10: 942. [42] Shao Y, Li M, Yu Q, et al. CircRNA CDR1as promotes cardiomyocyte apoptosis through activating hippo signaling pathway in diabetic cardiomyopathy. Eur J Pharmacol, 2022; 922: 174915. -


 投稿系统
投稿系统


 下载:
下载: